Prebiotic Factors Influencing the Activity of a Ligase Ribozyme
Abstract
:1. Introduction
2. Experimental Section
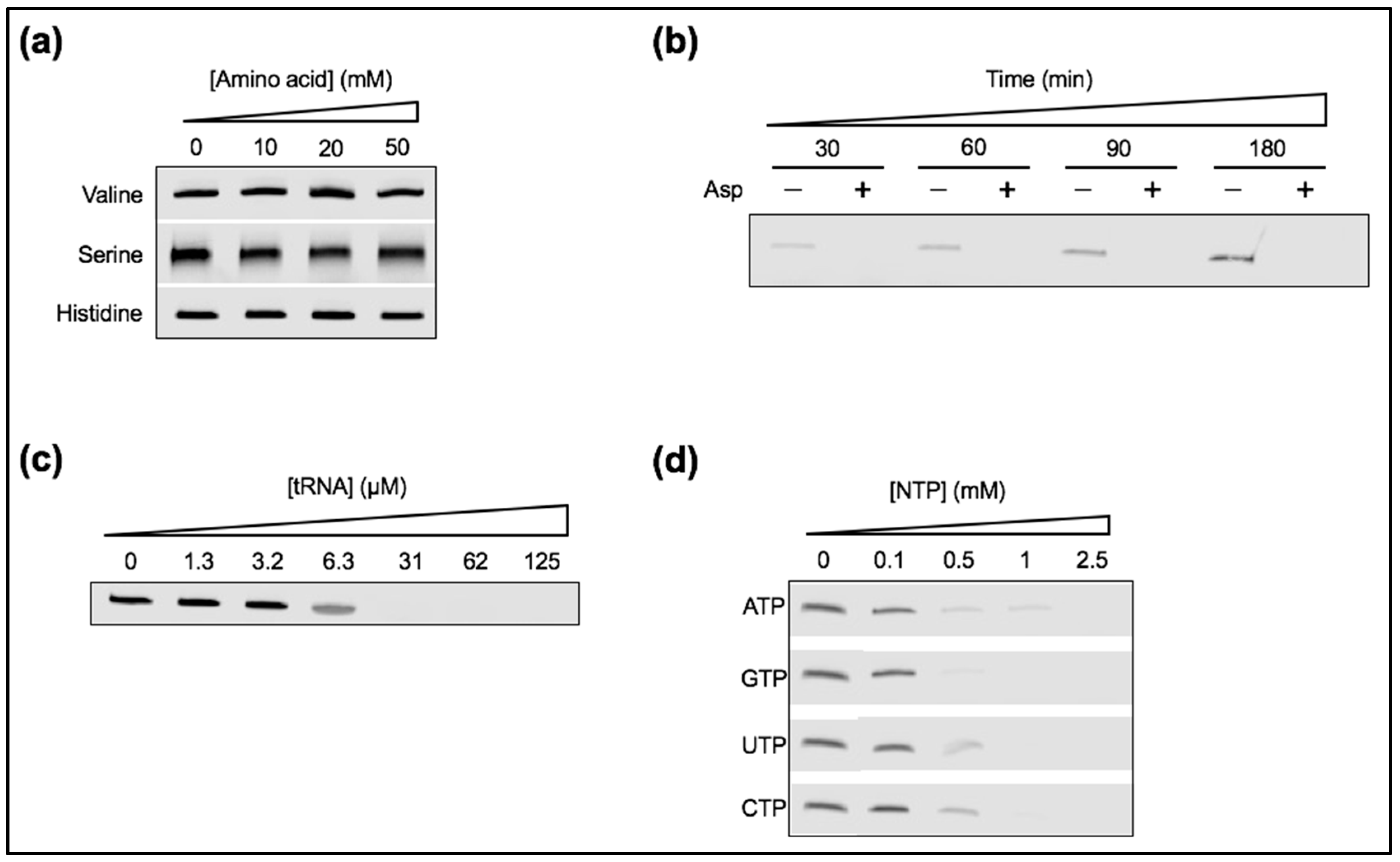
2.1. Materials
2.2. Preparation of Ribozyme
2.3. Ribozyme Activity Assay
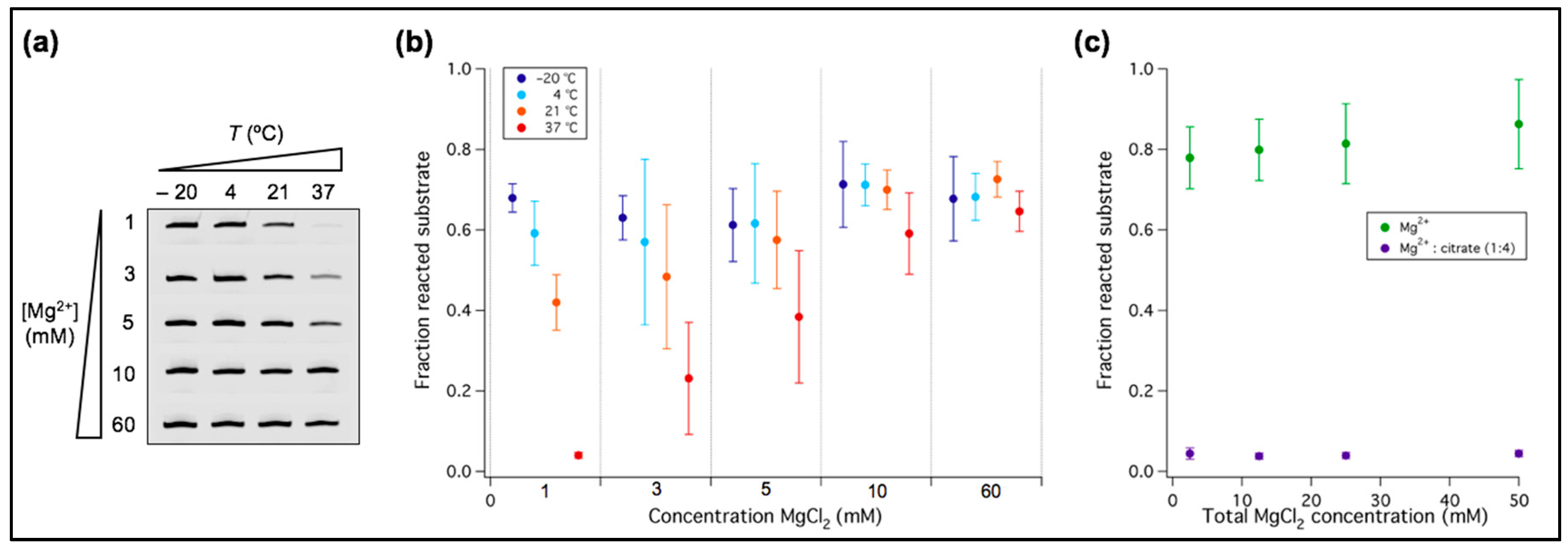
2.4. Ribozyme Activity Assay in the Presence of Citric Acid
2.5. Preparation of Micelles and Vesicles for Ribozyme Activity Assays
2.6. Ribozyme Activity Assay in the Presence of Fatty Acids
2.7. Ribozyme Activity Assay in the Presence of PEG
2.8. Ribozyme Activity Assay in the Presence of Various Solutes
2.9. Ribozyme Activity Assay in the Presence of a Mutated Substrate
2.10. Gel Analysis
2.11. Fluorescence Microscopy Imaging of Vesicles
3. Results and Discussion
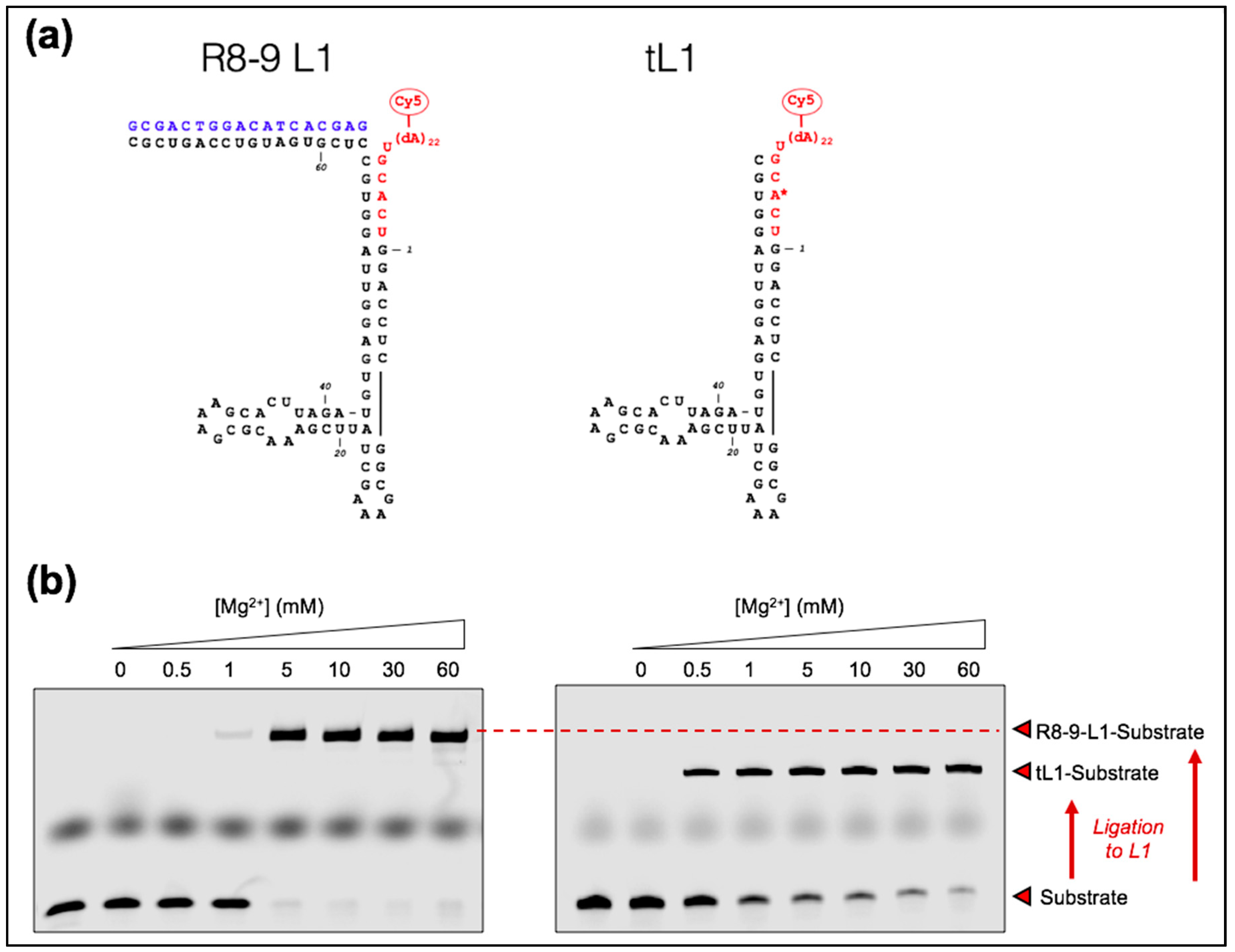
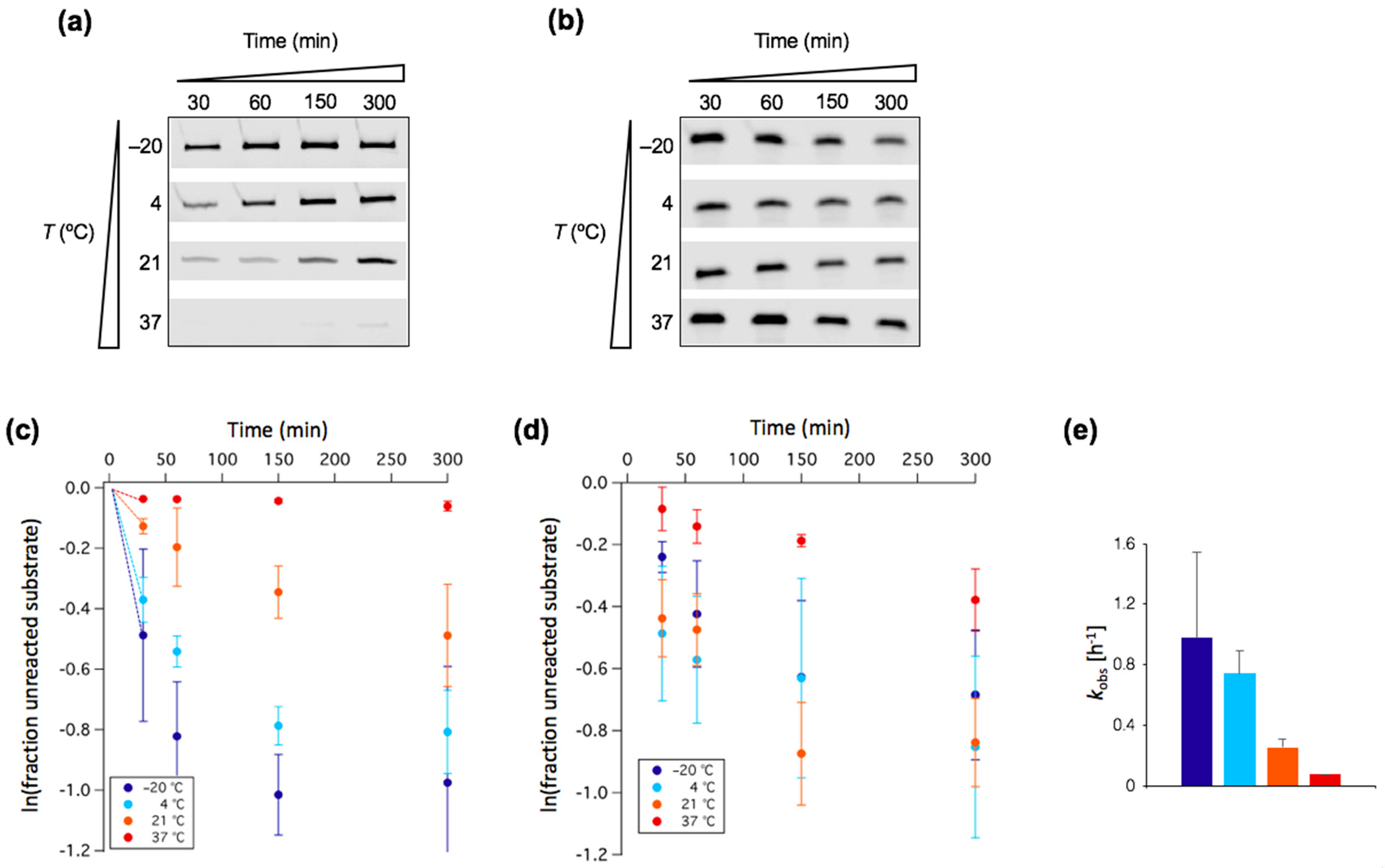
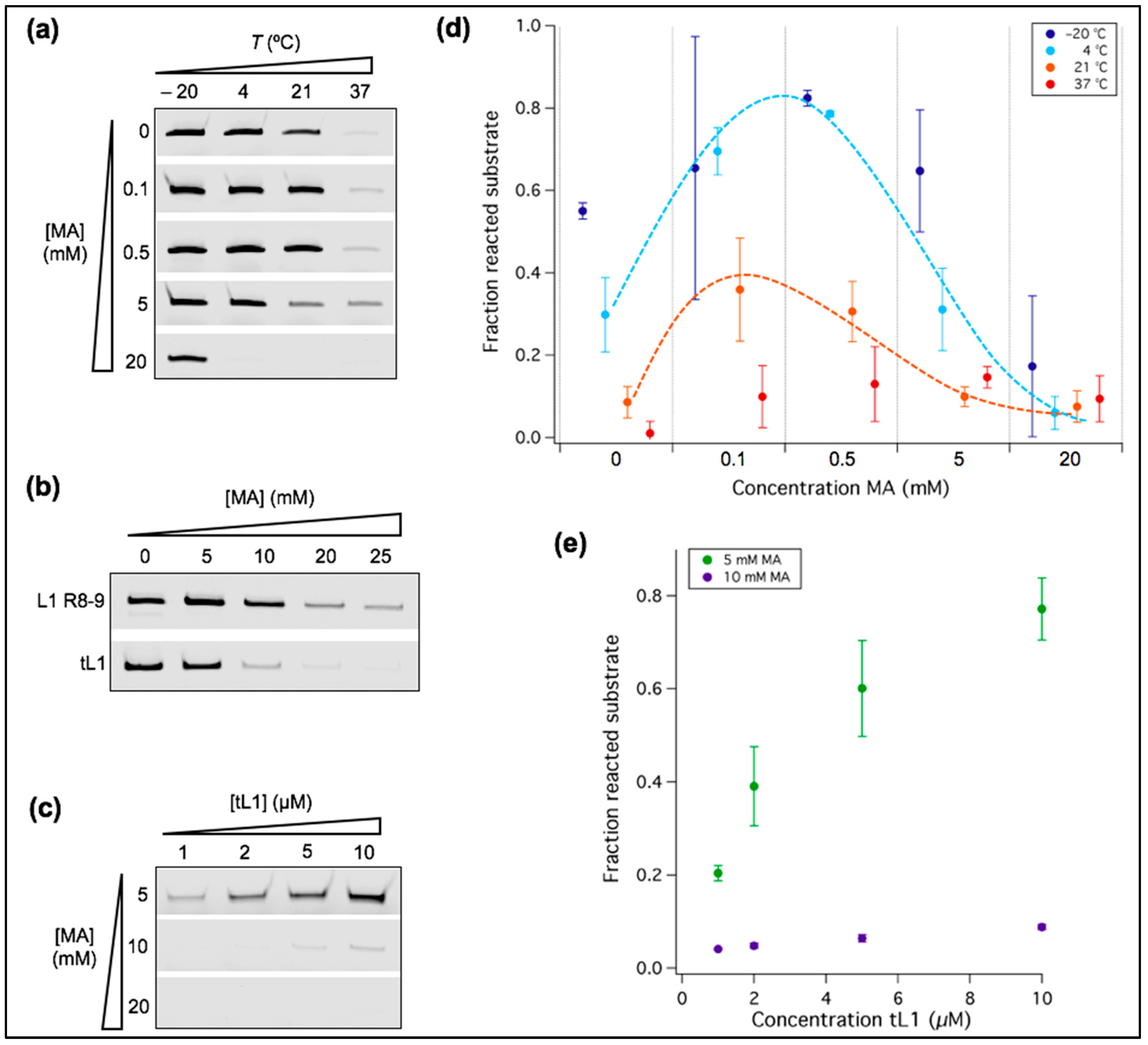
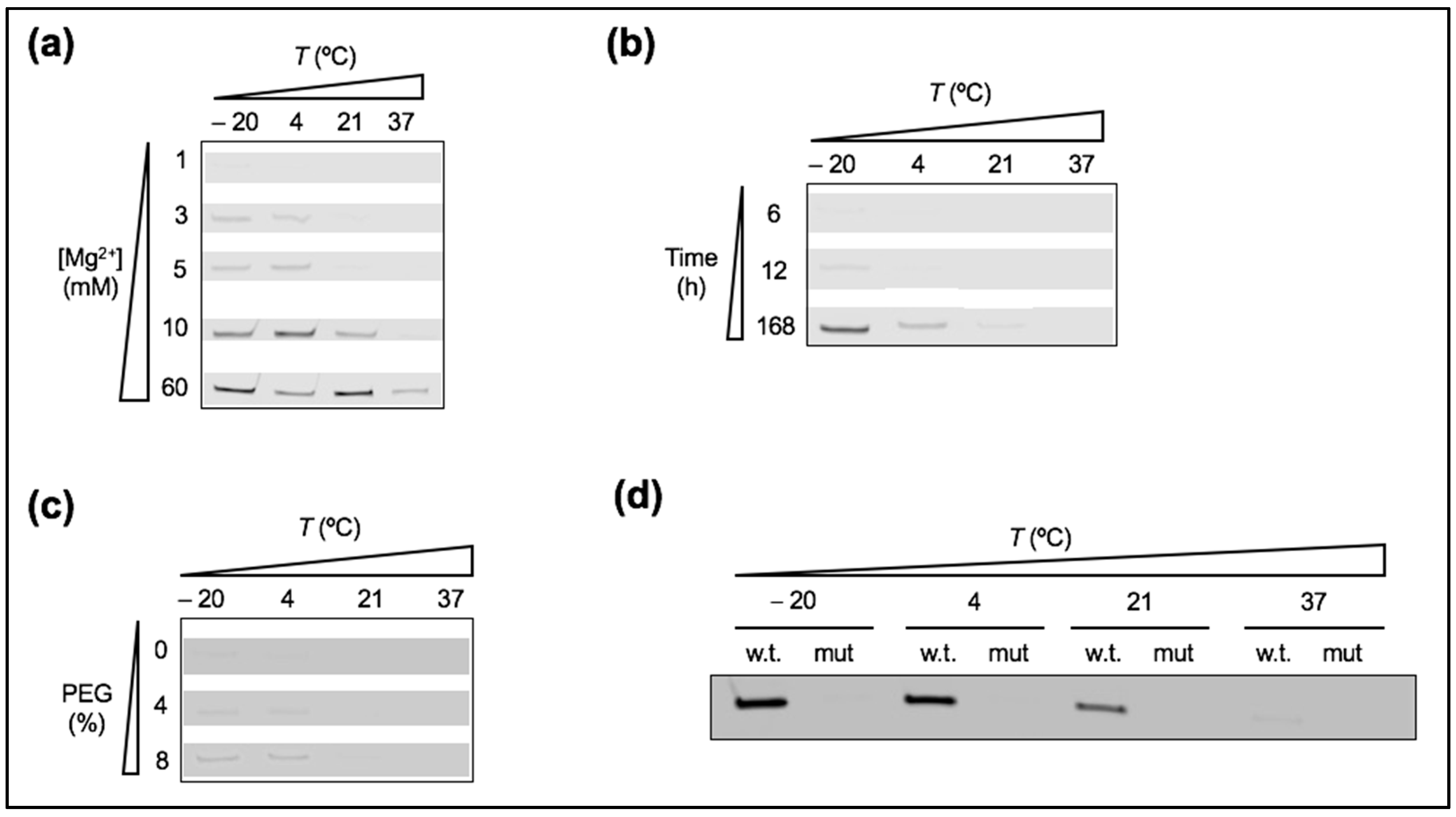
3.1. Low Temperature Decreases Mg2+ Requirement of a New Truncated L1 Ribozyme
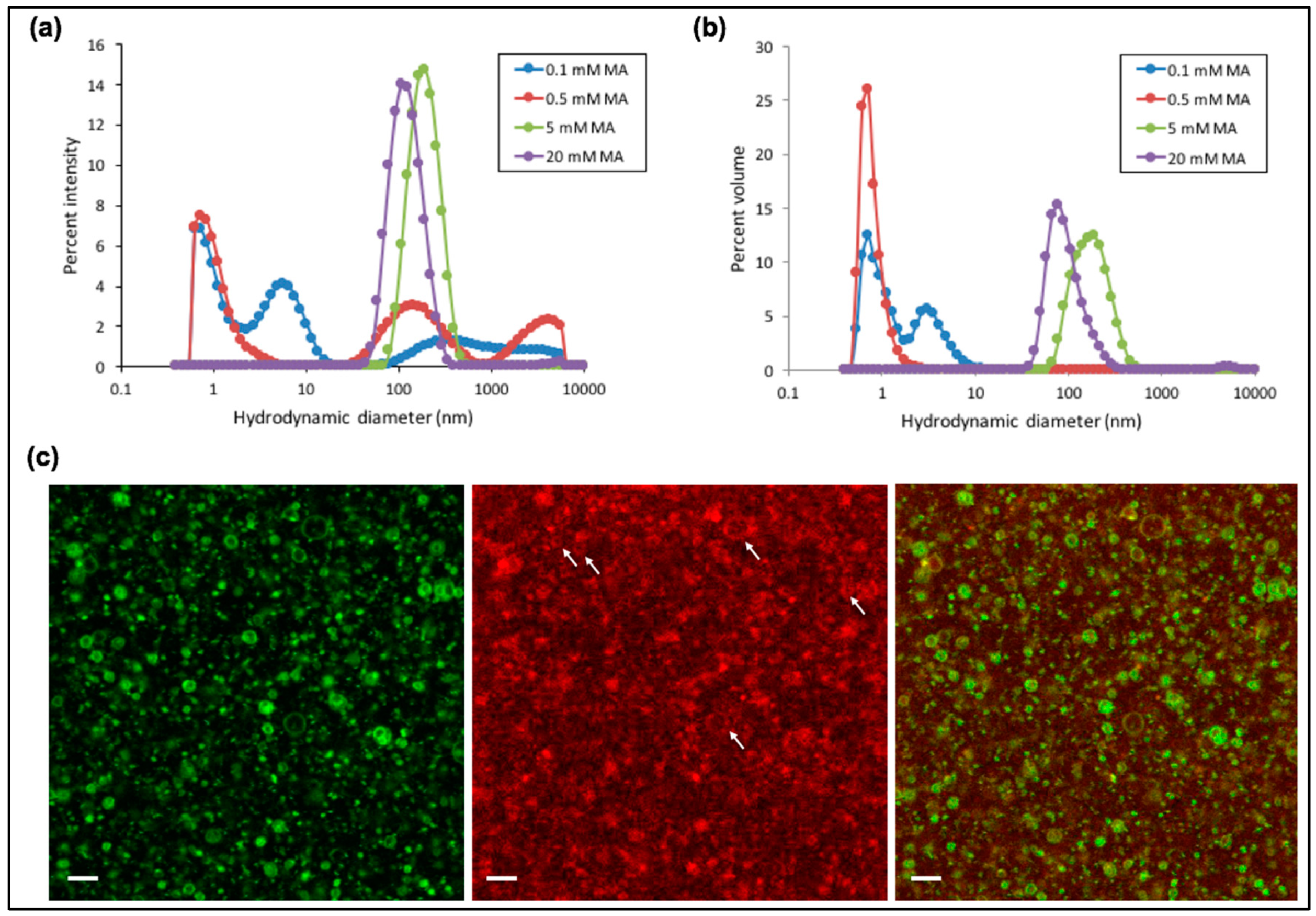
3.2. Characterization of Fatty Acid Micelles and Vesicles at 1 mM Mg2+
3.3. Ribozyme Activity Profile as a Function of Fatty Acid Concentration Exhibits a Maximum
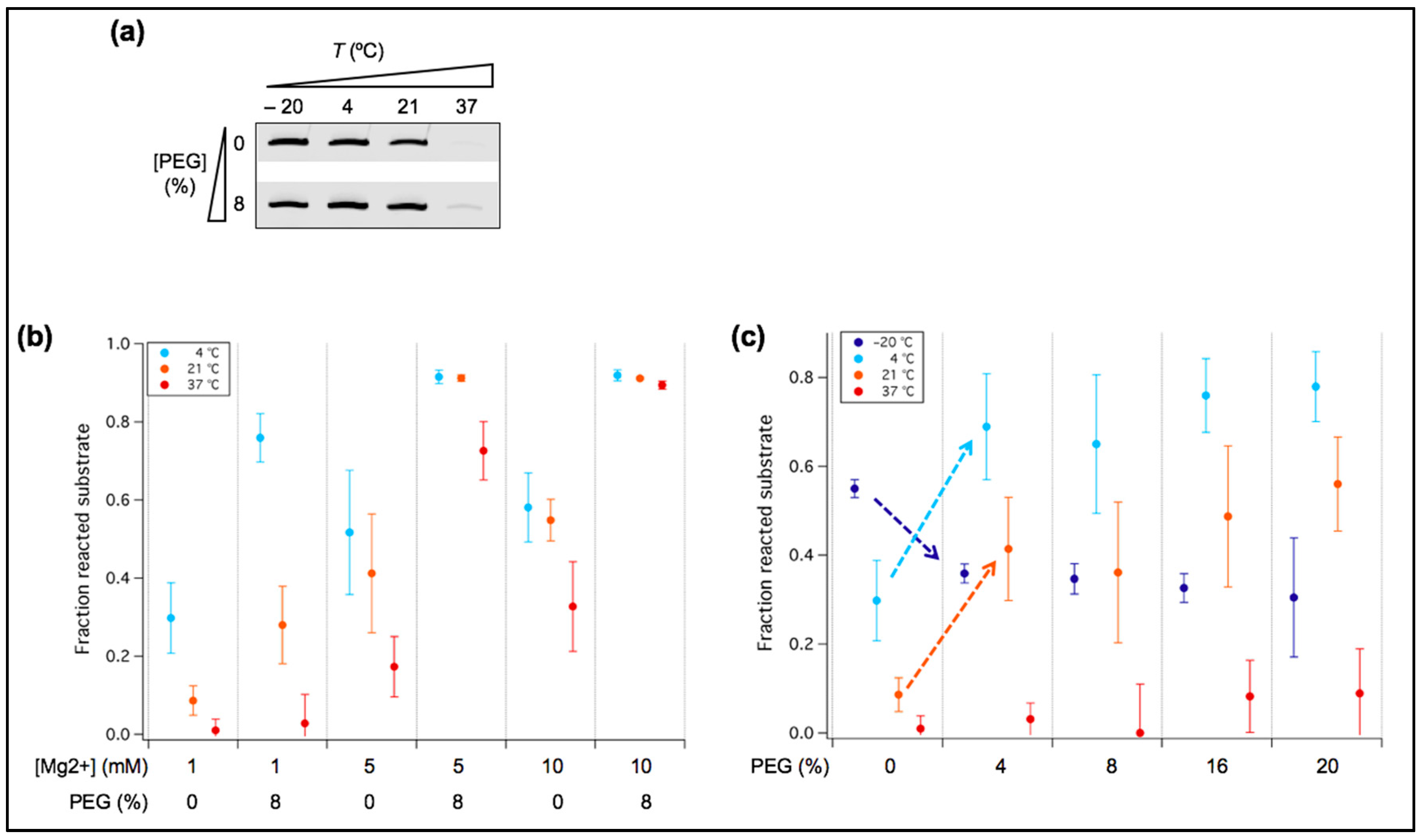
3.4. PEG Acts as a Positive or Negative Regulator of Ribozyme Activity Depending on Temperature
3.5. Lowering the Affinity between Ribozyme and Substrate Leads to Severe Loss of Reactivity
3.6. Ribozyme Activity Collapses in the Presence of Anionic Solutes
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Gilbert, W. The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Higgs, P.G.; Lehman, N. The RNA world: Molecular cooperation at the origin of life. Nat. Rev. Genet. 2015, 16, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Butlerow, A. Formation synthétique d’une substance sucrée. C. R. Acad. Sci. 1861, 53, 145–147. [Google Scholar]
- Patel, B.H.; Percivalle, C.; Ritson, D.J.; Duffy, C.D.; Sutherland, J.D. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 2015, 7, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Bad, J.L. How life began on Earth: A status report. Earth Planet. Sci. Lett. 2004, 226, 1–15. [Google Scholar] [CrossRef]
- Mutschler, H.; Lochner, A.; Holliger, P. Freeze-thaw cycles as drives of complex ribozyme assembly. Nat. Chem. 2015, 7, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Frommer, J.; Appel, B.; Müller, S. Ribozymes that can be regulated by external stimuli. Curr. Opin. Biotechnol. 2015, 31, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Johnston, B.H.; Landweber, L.F.; Kazakow, S.A. Ligation activity of fragmented ribozyme in frozen solution: Implication for the RNA world. Nucleic Acids Res. 2004, 32, 2966–2974. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Kazakov, S.A.; Johnston, B.H.; Landweber, L.F. The RNA world on ice: A new scenario for the emergence of RNA information. J. Mol. Evol. 2005, 61, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Szostak, J.W. The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 2012, 3, 2. [Google Scholar] [CrossRef]
- Inoue, A.; Takagi, Y.; Taira, K. Importance in catalysis of a magnesium ion with very low affinity for a hammerhead ribozyme. Nucleic Acids Rese. 2004, 32, 4217–4223. [Google Scholar] [CrossRef] [PubMed]
- Amotov, S.; Nishikawa, S.; Taira, K. Dependence on Mg2+ ions of the activities of dimeric hammerhead minizymes. FEBS Lett. 1996, 386, 99–102. [Google Scholar] [CrossRef]
- Anella, F.; Danelon, C. Reconciling ligase ribozyme activity with fatty acid vesicles stability. Life 2014, 4, 929–943. [Google Scholar] [CrossRef] [PubMed]
- Desai, R.; Kilburn, D.; Lee, H.T.; Woodson, S.A. Increased Ribozyme activity in crowded solutions. J. Biol. Chem. 2014, 289, 2972–2977. [Google Scholar] [CrossRef] [PubMed]
- Paddle, B.P.; Rueda, D. Molecular crowding accelerates ribozyme docking and catalysis. J. Am. Chem. Soc. 2014, 136, 16700–16703. [Google Scholar]
- Strulson, C.A.; Yennawar, N.H.; Rambo, R.P.; Bevilacqua, P.C. Molecular crowding favours reactivity of a human ribozyme under physiological ionic conditions. Biochemistry 2013, 52, 8187–8197. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Sung, H.L.; Nesbitt, D.J. Amino acid specific effects on RNA tertiary interactions: Single-molecule kinetic and thermodynamic studies. J. Phys. Chem. B 2016, 120, 10615–10627. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Kitigawa, Y.; Miyoshi, D.; Sugimoto, N. Effects of background anionic compounds on the activity of the hammerhead ribozyme in Mg2+-unsaturated solutions. J. Biol. Inorg. Chem. 2015, 20, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.K.; Draper, D.E. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 2002, 317, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.C.; Lenz, T.K.; Hud, N.V.; Williams, L.D. Cations in charge: Magnesium ions in RNA folding and catalysis. Curr. Opin. Struct. Biol. 2012, 22, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Hamper, A.; Cowan, J.A. A unique mechanism for RNA catalysis: The role of metal cofactor in hairpin ribozyme cleavage. Chem. Biol. 1997, 4, 513–517. [Google Scholar] [CrossRef]
- Hanna, R.; Doudna, J.A. Metal ions in ribozyme folding and catalysis. Curr. Opin. Chem. Biol. 2000, 4, 166–170. [Google Scholar] [CrossRef]
- Chen, I.A.; Walde, P. From self-assembled vesicles to protocells. Cold Spring Harbor Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Attwater, J.; Hollinger, P. A synthetic approach to abiogenesis. Nat. Methods 2014, 11, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Fujikawa, S.M.; Szostak, J.W. Experimental models of primitive cellular compartments: Encapsulation, growth and division. Science 2003, 302, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.A.; Salehi-Ashtiani, K.; Szostak, J.W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005, 127, 13213–13219. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Hesselberth, J.R.; Ellington, A.D. Optimization and optimality of a short ribozyme ligase that joins non-Watson-Crick base pairings. RNA 2001, 7, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Scott, W.G. The structural basis of ribozyme-catalyzed RNA assembly. Science 2007, 315, 1549–1553. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. A glimpse of biology's first enzyme. Science 2007, 315, 1507–1508. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Adamala, K.; Szostak, J.W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 2013, 342, 1098–1100. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, T.; Lehman, N. Non-unity molecular heritability demonstrated by continuous evolution in vitro. Chem. Biol. 1999, 6, 857–869. [Google Scholar] [CrossRef]
- Giambasu, G.M.; Lee, T.S.; Scott, W.G.; York, D.M. Mapping L1 Ligase ribozyme conformational switch. J. Mol. Biol. 2012, 423, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, S.E.; Balatskaya, S.V.; Johnston, B.H. Ligation of the hairpin ribozyme in cis induced by freezing and dehydration. RNA 2006, 12, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Hansma, H.G. The power of crowding for the origins of life. Orig. Life Evol. Biosph. 2014, 44, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Mast, C.B.; Schink, S.; Gerland, U.; Braun, D. Escalation of polymerization in a thermal gradient. Proc. Natl. Acad. Sci. USA 2013, 110, 8030–8035. [Google Scholar] [CrossRef] [PubMed]
- Glasner, M.E.; Bergman, N.H.; Bartel, D.P. Metal ion requirements for structure and catalysis of an RNA ligase ribozyme. Biochemistry 2002, 41, 8103–8112. [Google Scholar] [CrossRef] [PubMed]
- Mulkidjanian, A.Y.; Bychkov, A.Y.; Dibrova, D.V.; Galperin, M.Y.; Koonin, E.V. Origins of first cells at terrestrial, anoxic geothermal fields. Proc. Natl. Acad. Sci. USA 2012, 109, E821–E830. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anella, F.; Danelon, C. Prebiotic Factors Influencing the Activity of a Ligase Ribozyme. Life 2017, 7, 17. https://doi.org/10.3390/life7020017
Anella F, Danelon C. Prebiotic Factors Influencing the Activity of a Ligase Ribozyme. Life. 2017; 7(2):17. https://doi.org/10.3390/life7020017
Chicago/Turabian StyleAnella, Fabrizio, and Christophe Danelon. 2017. "Prebiotic Factors Influencing the Activity of a Ligase Ribozyme" Life 7, no. 2: 17. https://doi.org/10.3390/life7020017




