Characterization of Reconstructed Ancestral Proteins Suggests a Change in Temperature of the Ancient Biosphere
Abstract
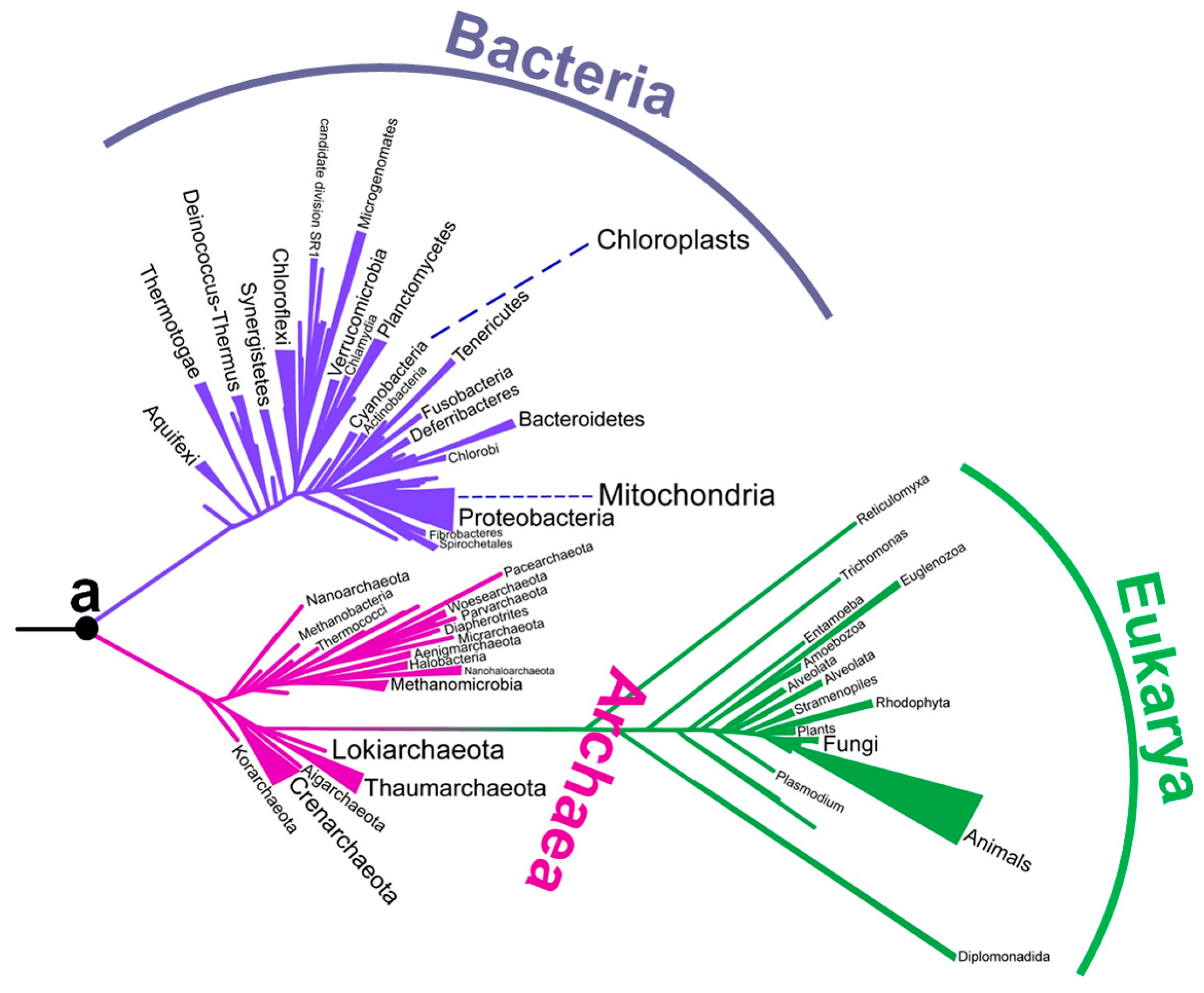
:1. Introduction
2. Early Studies on the Environmental Temperatures of Ancestral Life
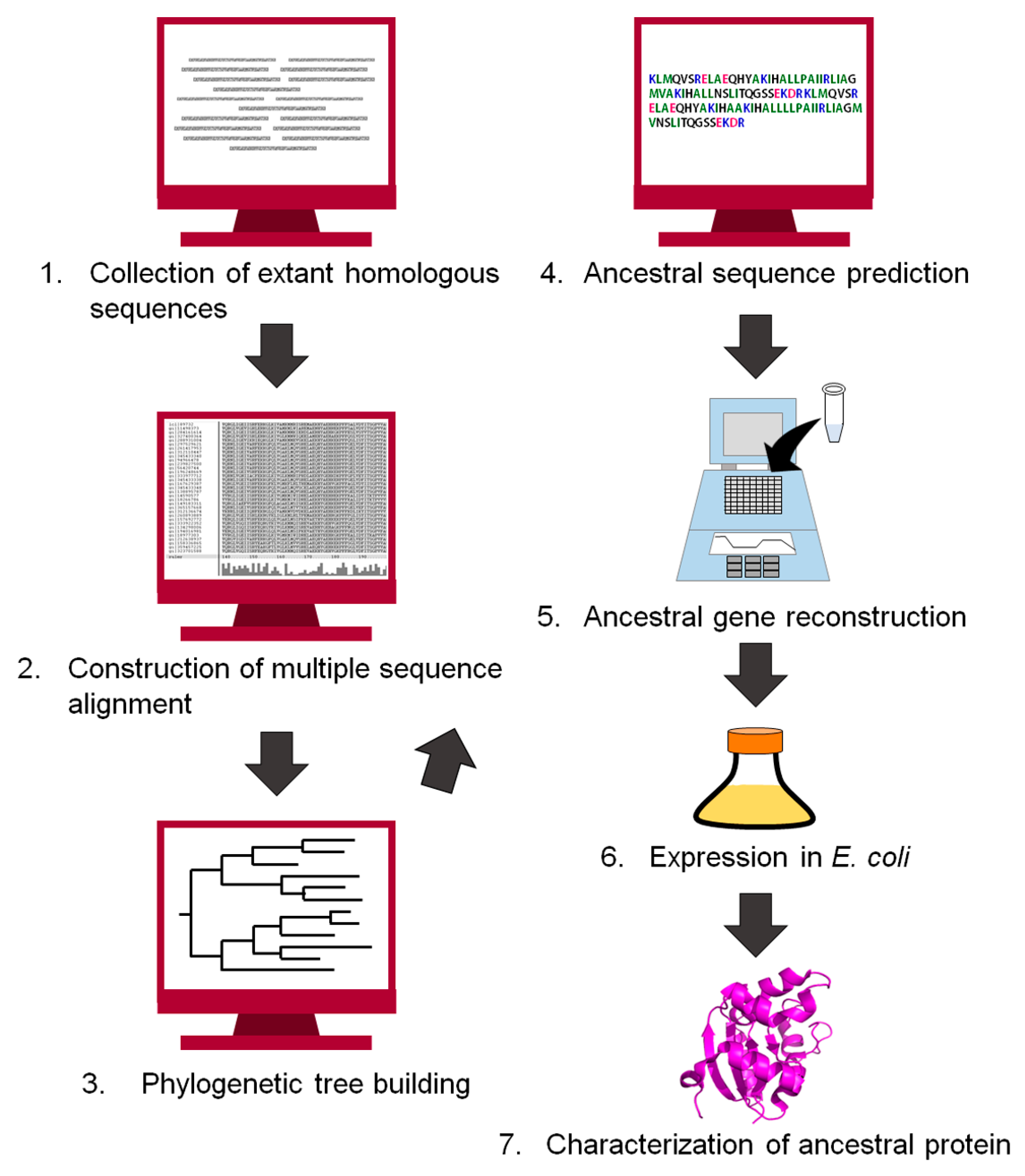
3. Experimental Procedure to Reconstruct an Ancestral Protein Sequence
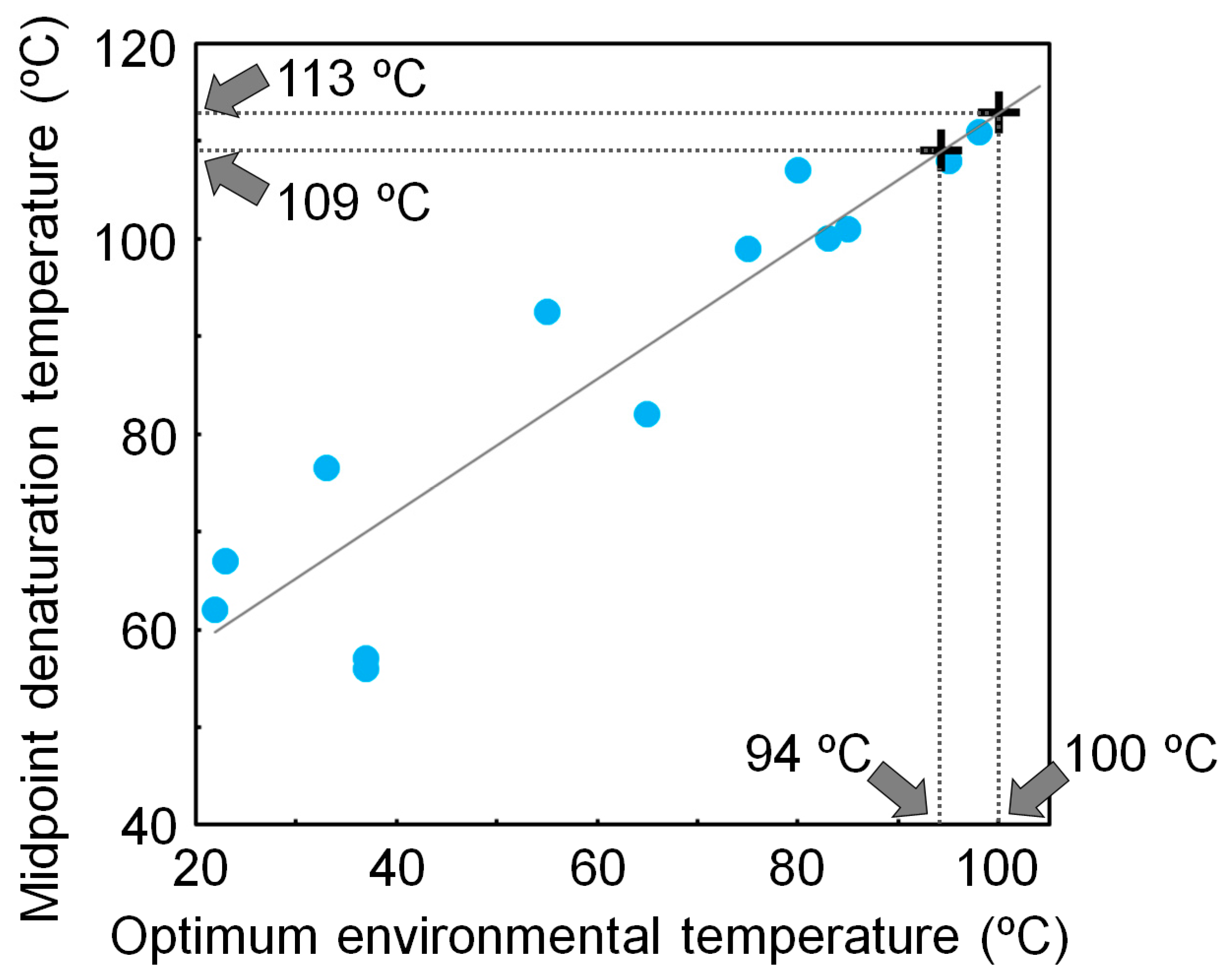
4. Experimentally Testing If Ancestral Organisms Were Thermophiles
5. Ancestral Sequence Reconstruction Using a Non-Homogeneous Model
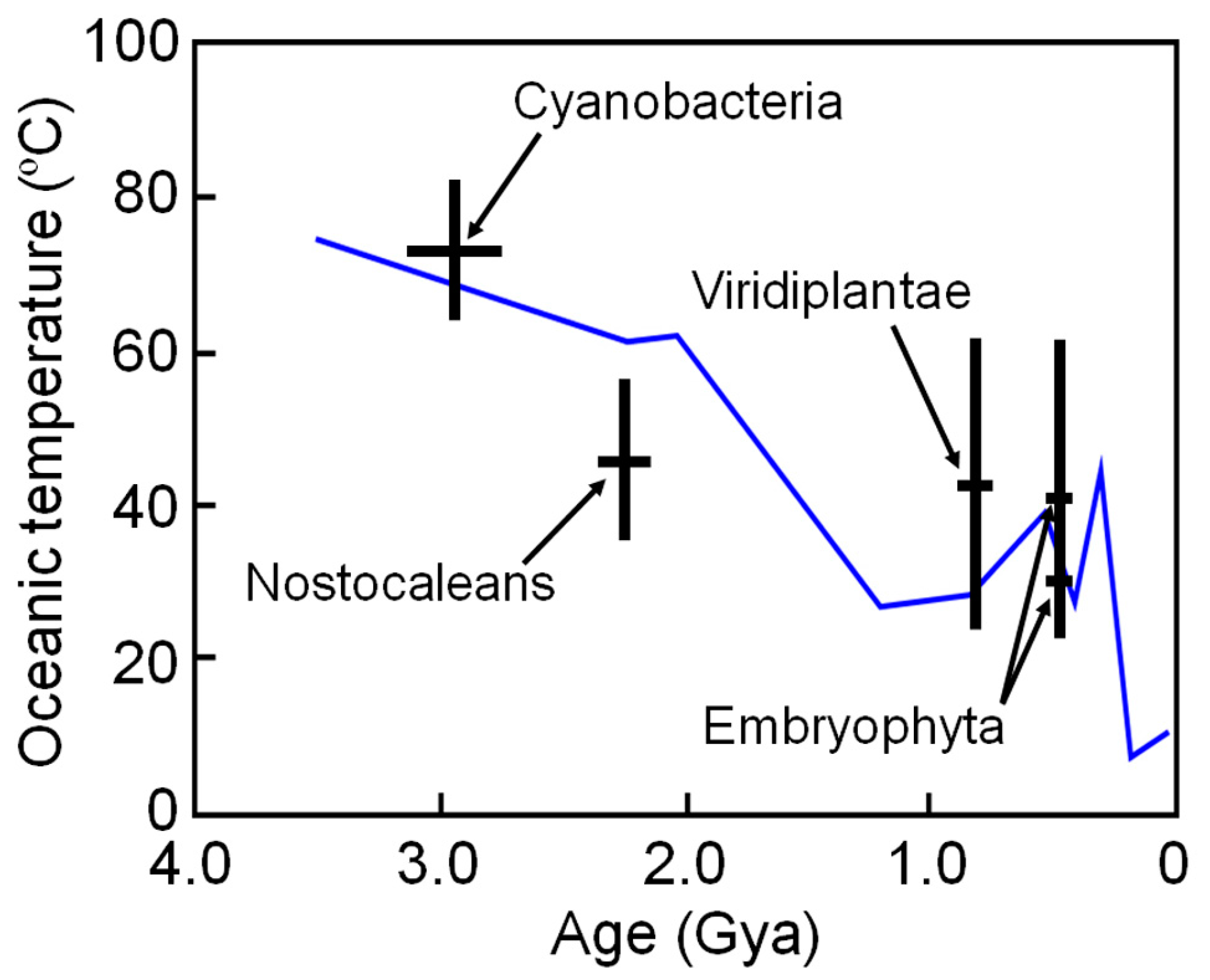
6. Estimating Long-Term Change in Biosphere Temperature
7. Limitations of Ancestral Inference to Estimate Temperatures of Ancient Biosphere
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Schopf, J.W. Microfossils of the early Archean apex chert: New evidence of the antiquity of life. Science 1993, 260, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Wacey, D.; McLoughlin, N.; Green, O.; Parnell, J.; Stoakes, C.A.; Brasier, M.D. The ~3.4 billion-year-old strelley pool sandstone: A new window into early life on Earth. Int. J. Astrobiol. 2006, 5, 333. [Google Scholar] [CrossRef]
- Nutman, A.P.; Bennett, V.C.; Friend, C.R.; Van Kranendonk, M.J.; Chivas, A.R. Rapid emergence of life shown by discovery of 3700-million-year-old microbial structures. Nature 2016, 537, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Dodd, M.S.; Papineau, D.; Grenne, T.; Slack, J.F.; Rittner, M.; Pirajno, F.; O’Neil, J.; Little, C.T. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nature 2017, 543, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Buick, R.; Canfield, D.E. Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature 2001, 410, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Ueno, Y.; Yamada, K.; Yoshida, N.; Maruyama, S.; Isozaki, Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 2006, 440, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Rosing, M.T. 13C-depleted carbon microparticles in >3700-Ma sea-floor sedimentary rocks from west Greenland. Science 1999, 283, 674–676. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.A.; Boehnke, P.; Harrison, T.M.; Mao, W.L. Potentially biogenic carbon preserved in a 4.1 billion-year-old zircon. Proc. Natl. Acad. Sci. USA 2015, 112, 14518–14521. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.W. Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat. Rev. Genet. 2004, 5, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, E.A.; Kratzer, J.T.; Randall, R.N. Deep phylogeny—How a tree can help characterize early life on Earth. Cold. Spring Harb. Perspect. Biol. 2010, 2, a002238. [Google Scholar] [CrossRef] [PubMed]
- Merkl, R.; Sterner, R. Ancestral protein reconstruction: Techniques and applications. Biol. Chem. 2016, 397, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Perez-Jimenez, R.; Ingles-Prieto, A.; Zhao, Z.M.; Sanchez-Romero, I.; Alegre-Cebollada, J.; Kosuri, P.; Garcia-Manyes, S.; Kappock, T.J.; Tanokura, M.; Holmgren, A.; et al. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat. Struct. Mol. Biol. 2011, 18, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Akanuma, S.; Yamagishi, A. A strategy for designing thermostable enzymes by reconstructing ancestral sequences possessed by ancient life. In Biotechnology of Extremophiles: Advances and Challenges; Rampelotto, P.H., Ed.; Springer: Berlin, Germany, 2016; pp. 581–596. [Google Scholar] [CrossRef]
- Boussau, B.; Gouy, M. What genomes have to say about the evolution of the earth. Gondwana Res. 2012, 21, 483–494. [Google Scholar] [CrossRef]
- Akanuma, S.; Nakajima, Y.; Yokobori, S.; Kimura, M.; Nemoto, N.; Mase, T.; Miyazono, K.; Tanokura, M.; Yamagishi, A. Experimental evidence for the thermophilicity of ancestral life. Proc. Natl. Acad. Sci. USA 2013, 110, 11067–11072. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, A.; Kon, T.; Takahashi, G.; Oshima, T. From the common ancestor of all living organisms to protoeukaryotic cell. In Thermophiles: The Keys to Molecular Evolution and the Origin of Life? Wiegel, J., Adams, M.W.W., Eds.; Taylor & Francis: Abingdon, UK, 1998; pp. 287–295. [Google Scholar]
- Akanuma, S.; Yokobori, S.; Nakajima, Y.; Bessho, M.; Yamagishi, A. Robustness of predictions of extremely thermally stable proteins in ancient organisms. Evolution 2015, 69, 2954–2962. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, R.; Nakagawa, M.; Kuroyanagi, T.; Yokobori, S.I.; Yamagishi, A. Quest for ancestors of eukaryal cells based on phylogenetic analyses of aminoacyl-tRNA synthetases. J. Mol. Evol. 2017, 84, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Pace, N.R. Origin of life—Facing up to the physical setting. Cell 1991, 65, 531–533. [Google Scholar] [CrossRef]
- Stetter, K.O. Hyperthermophilic procaryotes. FEMS Microbiol. Rev. 1996, 18, 149–158. [Google Scholar] [CrossRef]
- Brochier, C.; Philippe, H. Phylogeny: A non-hyperthermophilic ancestor for bacteria. Nature 2002, 417, 244. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Tourasse, N.; Gouy, M. A nonhyperthermophilic common ancestor to extant life forms. Science 1999, 283, 220–221. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. The universal ancestor lived in a thermophilic or hyperthermophilic environment. J. Theor. Biol. 2000, 203, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P. A hot topic: The origin of hyperthermophiles. Cell 1996, 85, 789–792. [Google Scholar] [CrossRef]
- Weiss, M.C.; Sousa, F.L.; Mrnjavac, N.; Neukirchen, S.; Roettger, M.; Nelson-Sathi, S.; Martin, W.F. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 2016, 1, 16116. [Google Scholar] [CrossRef] [PubMed]
- Boussau, B.; Blanquart, S.; Necsulea, A.; Lartillot, N.; Gouy, M. Parallel adaptations to high temperatures in the Archaean eon. Nature 2008, 456, 942–945. [Google Scholar] [CrossRef] [PubMed]
- Groussin, M.; Boussau, B.; Charles, S.; Blanquart, S.; Gouy, M. The molecular signal for the adaptation to cold temperature during early life on Earth. Biol. Lett. 2013, 9, 20130608. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [PubMed]
- Achenbach-Richter, L.; Gupta, R.; Zillig, W.; Woese, C.R. Rooting the archaebacterial tree: The pivotal role of Thermococcus celer in archaebacterial evolution. Syst. Appl. Microbiol. 1988, 10, 231–240. [Google Scholar] [CrossRef]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains archaea, bacteria, and eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L.; Lazcano, A. The origin of life—Did it occur at high temperatures? J. Mol. Evol. 1995, 41, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Larralde, R.; Robertson, M.P.; Miller, S.L. Rates of decomposition of ribose and other sugars: Implications for chemical evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 8158–8160. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, W.F. Phylogenetic classification and the universal tree. Science 1999, 284, 2124–2129. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. The universal ancestor and the ancestor of bacteria were hyperthermophiles. J. Mol. Evol. 2003, 57, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. The universal ancestor was a thermophile or a hyperthermophile: Tests and further evidence. J. Theor. Biol. 2003, 221, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P. A hot story from comparative genomics: Reverse gyrase is the only hyperthermophile-specific protein. Trends. Genet. 2002, 18, 236–237. [Google Scholar] [CrossRef]
- Atomi, H.; Matsumi, R.; Imanaka, T. Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 2004, 186, 4829–4833. [Google Scholar] [CrossRef] [PubMed]
- Declais, A.C.; Marsault, J.; Confalonieri, F.; de La Tour, C.B.; Duguet, M. Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J. Biol. Chem. 2000, 275, 19498–19504. [Google Scholar] [CrossRef] [PubMed]
- Heine, M.; Chandra, S.B. The linkage between reverse gyrase and hyperthermophiles: A review of their invariable association. J. Microbiol. 2009, 47, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Nisbet, E.G.; Sleep, N.H. The habitat and nature of early life. Nature 2001, 409, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, E.A.; Thomson, J.M.; Burgan, M.F.; Benner, S.A. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 2003, 425, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.M.; Gaucher, E.A.; Burgan, M.F.; De Kee, D.W.; Li, T.; Aris, J.P.; Benner, S.A. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat. Genet. 2005, 37, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, E.A.; Govindarajan, S.; Ganesh, O.K. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 2008, 451, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Butzin, N.C.; Lapierre, P.; Green, A.G.; Swithers, K.S.; Gogarten, J.P.; Noll, K.M. Reconstructed ancestral Myo-inositol-3-phosphate synthases indicate that ancestors of the Thermococcales and Thermotoga species were more thermophilic than their descendants. PLoS ONE 2013, 8, e84300. [Google Scholar] [CrossRef] [PubMed]
- Hart, K.M.; Harms, M.J.; Schmidt, B.H.; Elya, C.; Thornton, J.W.; Marqusee, S. Thermodynamic system drift in protein evolution. PLoS Biol. 2014, 12, e1001994. [Google Scholar] [CrossRef] [PubMed]
- Fournier, G.P.; Alm, E.J. Ancestral reconstruction of a pre-LUCA aminoacyl-tRNA synthetase ancestor supports the late addition of Trp to the genetic code. J. Mol. Evol. 2015, 80, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Busch, F.; Rajendran, C.; Heyn, K.; Schlee, S.; Merkl, R.; Sterner, R. Ancestral tryptophan synthase reveals functional sophistication of primordial enzyme complexes. Cell Chem. Biol. 2016, 23, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.K.; Schopf, J.W.; Yokobori, S.I.; Akanuma, S.; Yamagishi, A. Reconstructed ancestral enzymes suggest long-term cooling of Earth’s photic zone since the Archean. Proc. Natl. Acad. Sci. USA 2017, 114, 4619–4624. [Google Scholar] [CrossRef] [PubMed]
- Messier, W.; Stewart, C.B. Episodic adaptive evolution of primate lysozymes. Nature 1997, 385, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Bosnali, M.; Carstensen, L.; Seitz, T.; Durchschlag, H.; Blanquart, S.; Merkl, R.; Sterner, R. Computational and experimental evidence for the evolution of a (βα)8-barrel protein from an ancestral quarter-barrel stabilised by disulfide bonds. J. Mol. Biol. 2010, 398, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Paml: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997, 13, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Blanquart, S.; Lartillot, N. A site- and time-heterogeneous model of amino acid replacement. Mol. Biol. Evol. 2008, 25, 842–858. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.J.; Shields, D.C. GASP: Gapped ancestral sequence prediction for proteins. BMC Bioinform. 2004, 5, 123. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, J.; Nakaya, S.; Suzuki, T.; Tamakoshi, M.; Oshima, T.; Yamagishi, A. Ancestral residues stabilizing 3-isopropylmalate dehydrogenase of an extreme thermophile: Experimental evidence supporting the thermophilic common ancestor hypothesis. J. Biochem. 2001, 129, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Iwabata, H.; Watanabe, K.; Ohkuri, T.; Yokobori, S.; Yamagishi, A. Thermostability of ancestral mutants of Caldococcus noboribetus isocitrate dehydrogenase. FEMS Microbiol. Lett. 2005, 243, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Yokobori, S.; Ohkuri, T.; Yokogawa, T.; Nishikawa, K.; Yamagishi, A. Extremely thermophilic translation system in the common ancestor commonote: Ancestral mutants of glycyl-tRNA synthetase from the extreme thermophile Thermus thermophilus. J. Mol. Biol. 2007, 369, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Ohkuri, T.; Yokobori, S.; Yamagishi, A. Designing thermostable proteins: Ancestral mutants of 3-isopropylmalate dehydrogenase designed by using a phylogenetic tree. J. Mol. Biol. 2006, 355, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, K.; Yokobori, S.; Koikeda, S.; Yamagishi, A. Improvement of Bacillus circulans beta-amylase activity attained using the ancestral mutation method. Protein Eng. Des. Sel. 2010, 23, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Semba, Y.; Ishida, M.; Yokobori, S.; Yamagishi, A. Ancestral amino acid substitution improves the thermal stability of recombinant lignin-peroxidase from white-rot fungi, Phanerochaete chrysosporium strain UAMH 3641. Protein Eng. Des. Sel. 2015, 28, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Abe, A.; Tamura, T.; Kishimoto, T.; Sogabe, A.; Akanuma, S.; Yokobori, S.; Yamagishi, A.; Imada, K.; Inagaki, K. Epistasis effects of multiple ancestral-consensus amino acid substitutions on the thermal stability of glycerol kinase from Cellulomonas sp. NT3060. J. Biosci. Bioeng. 2016, 121, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.D.; Pollock, D.D.; Blackburne, B.P.; Goldstein, R.A. Assessing the accuracy of ancestral protein reconstruction methods. PLoS. Comput. Biol. 2006, 2, e69. [Google Scholar] [CrossRef] [PubMed]
- Bridgham, J.T.; Carroll, S.M.; Thornton, J.W. Evolution of hormone-receptor complexity by molecular exploitation. Science 2006, 312, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Ortlund, E.A.; Bridgham, J.T.; Redinbo, M.R.; Thornton, J.W. Crystal structure of an ancient protein: Evolution by conformational epistasis. Science 2007, 317, 1544–1548. [Google Scholar] [CrossRef] [PubMed]
- Bridgham, J.T.; Ortlund, E.A.; Thornton, J.W. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 2009, 461, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.M.; Ortlund, E.A.; Thornton, J.W. Mechanisms for the evolution of a derived function in the ancestral glucocorticoid receptor. PLoS Genet. 2011, 7, e1002117. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, G.C.; Hanson-Smith, V.; Stevens, T.H.; Thornton, J.W. Evolution of increased complexity in a molecular machine. Nature 2012, 481, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.K.; Kelley, S.T.; Spiegelman, G.B.; Pace, N.R. The genetic core of the universal ancestor. Genome Res. 2003, 13, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Ciccarelli, F.D.; Doerks, T.; von Mering, C.; Creevey, C.J.; Snel, B.; Bork, P. Toward automatic reconstruction of a highly resolved tree of life. Science 2006, 311, 1283–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yutin, N.; Makarova, K.S.; Mekhedov, S.L.; Wolf, Y.I.; Koonin, E.V. The deep archaeal roots of eukaryotes. Mol. Biol. Evol. 2008, 25, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M.C.; Lake, J.A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 1992, 257, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.J.; Foster, P.G.; Hirt, R.P.; Harris, S.R.; Embley, T.M. The archaebacterial origin of eukaryotes. Proc. Natl. Acad. Sci. USA 2008, 105, 20356–20361. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.A.; Foster, P.G.; Cox, C.J.; Embley, T.M. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 2013, 504, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Brochier-Armanet, C.; Gribaldo, S. The two-domain tree of life is linked to a new root for the archaea. Proc. Natl. Acad. Sci. USA 2015, 112, 6670–6675. [Google Scholar] [CrossRef] [PubMed]
- Eick, G.N.; Bridgham, J.T.; Anderson, D.P.; Harms, M.J.; Thornton, J.W. Robustness of reconstructed ancestral protein functions to statistical uncertainty. Mol. Biol. Evol. 2017, 34, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Holinski, A.; Heyn, K.; Merkl, R.; Sterner, R. Combining ancestral sequence reconstruction with protein design to identify an interface hotspot in a key metabolic enzyme complex. Proteins 2017, 85, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.J.; Fresco, J.R.; Singh, M. A novel method for estimating ancestral amino acid composition and its application to proteins of the last universal ancestor. Bioinformatics 2004, 20, 2251–2257. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T. A co-evolution theory of the genetic code. Proc. Natl. Acad. Sci. USA 1975, 72, 1909–1912. [Google Scholar] [CrossRef] [PubMed]
- Baumann, U.; Oro, J. Three stages in the evolution of the genetic code. BioSystems 1993, 29, 133–141. [Google Scholar] [CrossRef]
- Trifonov, E.N. Consensus temporal order of amino acids and evolution of the triplet code. Gene 2000, 261, 139–151. [Google Scholar] [CrossRef]
- Akanuma, S.; Kigawa, T.; Yokoyama, S. Combinatorial mutagenesis to restrict amino acid usage in an enzyme to a reduced set. Proc. Natl. Acad. Sci. USA 2002, 99, 13549–13553. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.J.; Fresco, J.R.; Lesk, A.M.; Singh, M. Evolution of amino acid frequencies in proteins over deep time: Inferred order of introduction of amino acids into the genetic code. Mol. Biol. Evol. 2002, 19, 1645–1655. [Google Scholar] [CrossRef] [PubMed]
- Jordan, I.K.; Kondrashov, F.A.; Adzhubei, I.A.; Wolf, Y.I.; Koonin, E.V.; Kondrashov, A.S.; Sunyaev, S. A universal trend of amino acid gain and loss in protein evolution. Nature 2005, 433, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.M.; Lee, J.; Blaber, M. Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein. Proc. Natl. Acad. Sci. USA 2013, 110, 2135–2139. [Google Scholar] [CrossRef] [PubMed]
- Knauth, L.P.; Lowe, D.R. Oxygen isotope geochemistry of cherts from the Onverwacht Group (3.4 billion years), Transvaal, South Africa, with implications for secular variations in the isotopic composition of cherts. Earth Planet. Sci. Lett. 1978, 41, 209–222. [Google Scholar] [CrossRef]
- Robert, F.; Chaussidon, M. A palaeotemperature curve for the Precambrian oceans based on silicon isotopes in cherts. Nature 2006, 443, 969–972. [Google Scholar] [CrossRef] [PubMed]
- Groussin, M.; Gouy, M. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Mol. Biol. Evol. 2011, 28, 2661–2674. [Google Scholar] [CrossRef] [PubMed]
- Steipe, B.; Schiller, B.; Pluckthun, A.; Steinbacher, S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 1994, 240, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Davidson, A.R. The design of a hyperstable mutant of the Abp1p SH3 domain by sequence alignment analysis. Protein Sci. 2000, 9, 2457–2469. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Kostrewa, D.; Wyss, M.; Brugger, R.; D’Arcy, A.; Pasamontes, L.; van Loon, A.P. From DNA sequence to improved functionality: Using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng. 2000, 13, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Steipe, B. Consensus-based engineering of protein stability: From intrabodies to thermostable enzymes. Methods Enzymol. 2004, 388, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Loening, A.M.; Fenn, T.D.; Wu, A.M.; Gambhir, S.S. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 2006, 19, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, M.; Tawfik, D.S. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc. Natl. Acad. Sci. USA 2009, 106, 6197–6202. [Google Scholar] [CrossRef] [PubMed]
- Trudeau, D.L.; Kaltenbach, M.; Tawfik, D.S. On the potential origins of the high stability of reconstructed ancestral proteins. Mol. Biol. Evol. 2016, 33, 2633–2641. [Google Scholar] [CrossRef] [PubMed]
- Akanuma, S.; Yokobori, S.; Yamagishi, A. Comparative genomics of thermophilic bacteria and archaea. In Thermophilic Microbes in Environmental and Industrial Biotechnology; Satyanarayana, T., Litterchild, J., Kawarabayasi, Y., Eds.; Springer: Berlin, Germany, 2013; pp. 331–349. [Google Scholar]
- Iwabe, N.; Kuma, K.; Hasegawa, M.; Osawa, S.; Miyata, T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl. Acad. Sci. USA 1989, 86, 9355–9359. [Google Scholar] [CrossRef] [PubMed]
- Gogarten, J.P.; Kibak, H.; Dittrich, P.; Taiz, L.; Bowman, E.J.; Bowman, B.J.; Manolson, M.F.; Poole, R.J.; Date, T.; Oshima, T.; et al. Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. Proc. Natl. Acad. Sci. USA 1989, 86, 6661–6665. [Google Scholar] [PubMed]
- Brown, J.R.; Doolittle, W.F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl. Acad. Sci. USA 1995, 92, 2441–2445. [Google Scholar] [CrossRef] [PubMed]
- Fournier, G.P.; Andam, C.P.; Alm, E.J.; Gogarten, J.P. Molecular evolution of aminoacyl tRNA synthetase proteins in the early history of life. Orig. Life Evol. Biosph. 2011, 41, 621–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalier-Smith, T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 2002, 52, 7–76. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Cell evolution and Earth history: Stasis and revolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 969–1006. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Rooting the tree of life by transition analyses. Biol. Direct 2006, 1, 19. [Google Scholar] [CrossRef] [PubMed]
- Lake, J.A.; Servin, J.A.; Herbold, C.W.; Skophammer, R.G. Evidence for a new root of the tree of life. Syst. Biol. 2008, 57, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Lake, J.A.; Skophammer, R.G.; Herbold, C.W.; Servin, J.A. Genome beginnings: Rooting the tree of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Deep phylogeny, ancestral groups and the four ages of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 111–132. [Google Scholar] [CrossRef] [PubMed]
- Groussin, M.; Hobbs, J.K.; Szollosi, G.J.; Gribaldo, S.; Arcus, V.L.; Gouy, M. Toward more accurate ancestral protein genotype-phenotype reconstructions with the use of species tree-aware gene trees. Mol. Biol. Evol. 2015, 32, 13–22. [Google Scholar] [CrossRef] [PubMed]
| Target | Method | Conclusion | Refs. |
|---|---|---|---|
| Common ancestors of Archaea and Bacteria | rRNA tree | Hyperthermophilic | [19,20] |
| Bacterial common ancestors | rRNA tree | Mesophilic or thermophilic | [21] |
| LUCA | G + C content in rRNA | Mesophilic | [22] |
| LUCA | Reanalysis of the data used in [22] | Thermophilic or hyperthermophilic | [23] |
| LUCA | Evolution of reverse gyrase | Mesophilic or thermophilic | [24] |
| LUCA | A gene for reverse gyrase found in a gene set of LUCA | Hyperthermophilic | [25] |
| LUCA | G + C contents in rRNA and amino acid composition inferred using a non-homogeneous model | Psychrophilic or mesophilic | [26] |
| LUCA | Amino acid composition inferred using a non-homogeneous model | Mesophilic | [27] |
| Target | Method | Conclusion | Refs. |
|---|---|---|---|
| LUCA | Introduction of a few amino acids into the sequence of a modern thermophilic protein | Hyperthermophilic | [55,56,57,58] |
| Bacterial common ancestors | Reconstruction of ancestral elongation factors | Thermophilic | [41,43] |
| Common ancestor of Thermotogales | Reconstruction of ancestral Myo-inositol-3-phospate synthases | Hyperthermophilic | [44] |
| LUCA | Reconstruction of ancestral NDKs using a homogeneous substitution model | Thermophilic or hyperthermophilic | [15] |
| LUCA | Reconstruction of ancestral NDKs using a non-homogeneous substitution model | Hyperthermophilic | [17] |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akanuma, S. Characterization of Reconstructed Ancestral Proteins Suggests a Change in Temperature of the Ancient Biosphere. Life 2017, 7, 33. https://doi.org/10.3390/life7030033
Akanuma S. Characterization of Reconstructed Ancestral Proteins Suggests a Change in Temperature of the Ancient Biosphere. Life. 2017; 7(3):33. https://doi.org/10.3390/life7030033
Chicago/Turabian StyleAkanuma, Satoshi. 2017. "Characterization of Reconstructed Ancestral Proteins Suggests a Change in Temperature of the Ancient Biosphere" Life 7, no. 3: 33. https://doi.org/10.3390/life7030033





