Gold Nanoparticles for Diagnostics: Advances towards Points of Care
Abstract
:1. Introduction
1.1. Gold Nanoparticles (AuNPs)—Properties and Sensing Applications
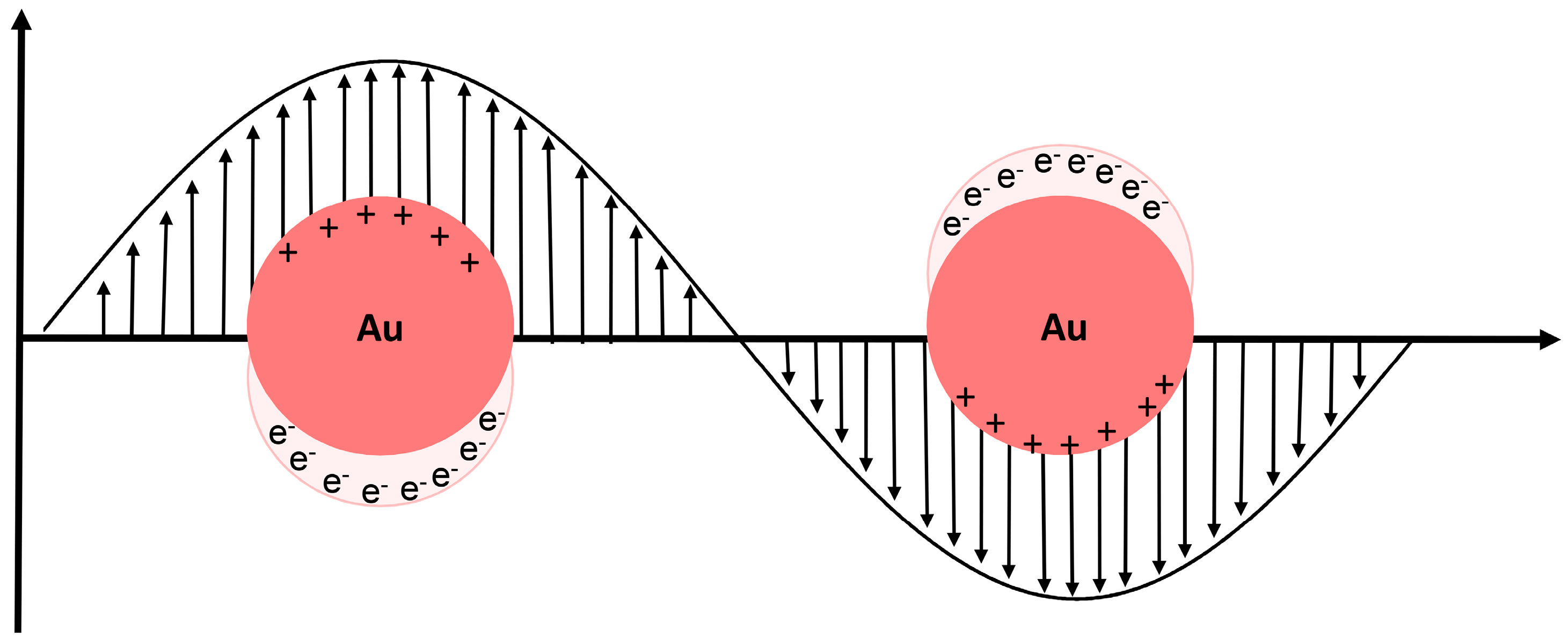
1.1.1. Localized Surface Plasmon Resonance (LSPR)
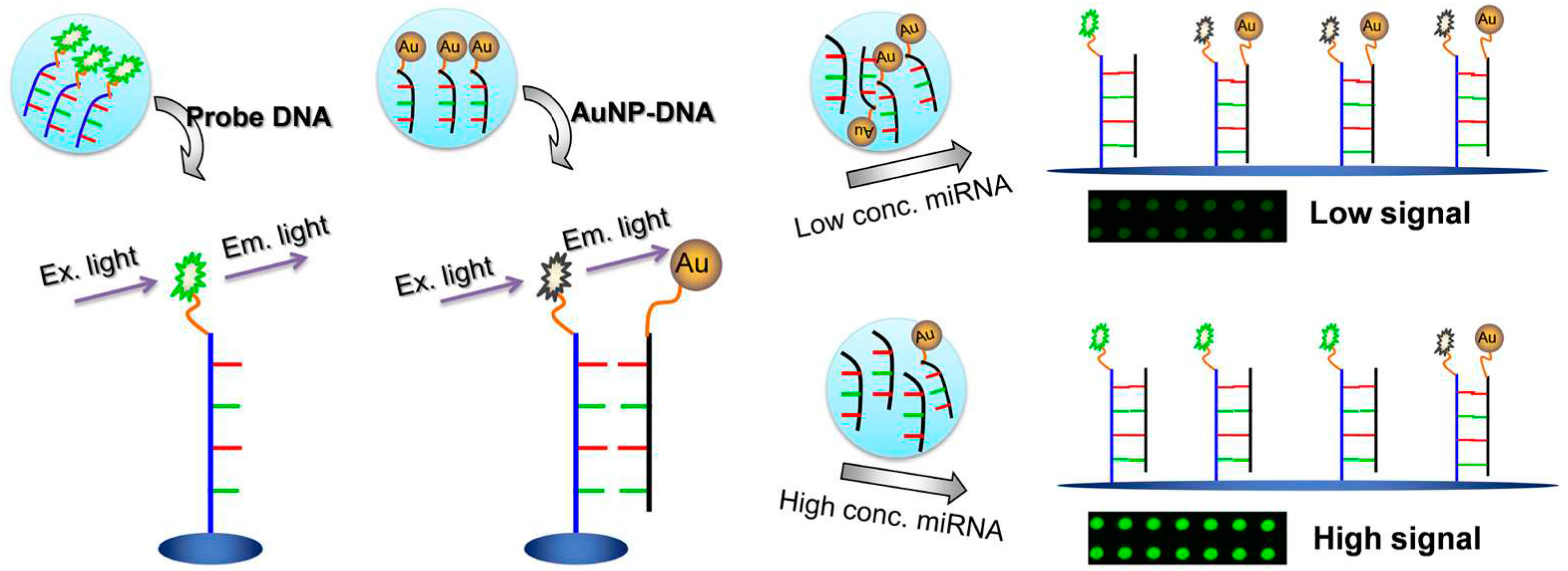
1.1.2. Fluorescence Modulation
1.1.3. Surface-Enhanced Raman Scattering (SERS)
1.1.4. Electrochemistry
2. General Principles of AuNP-Based Biomolecular Recognition
2.1. Nucleic Acids Sensing
2.2. Protein Sensing
3. General Overview of Applications
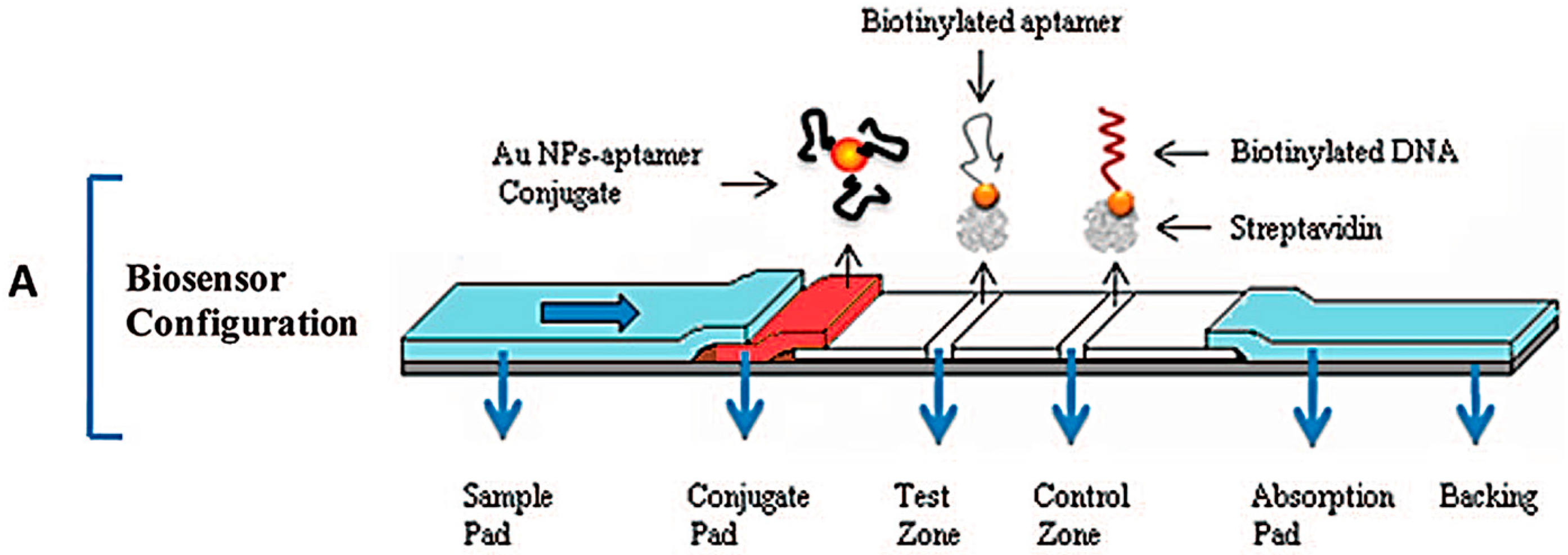
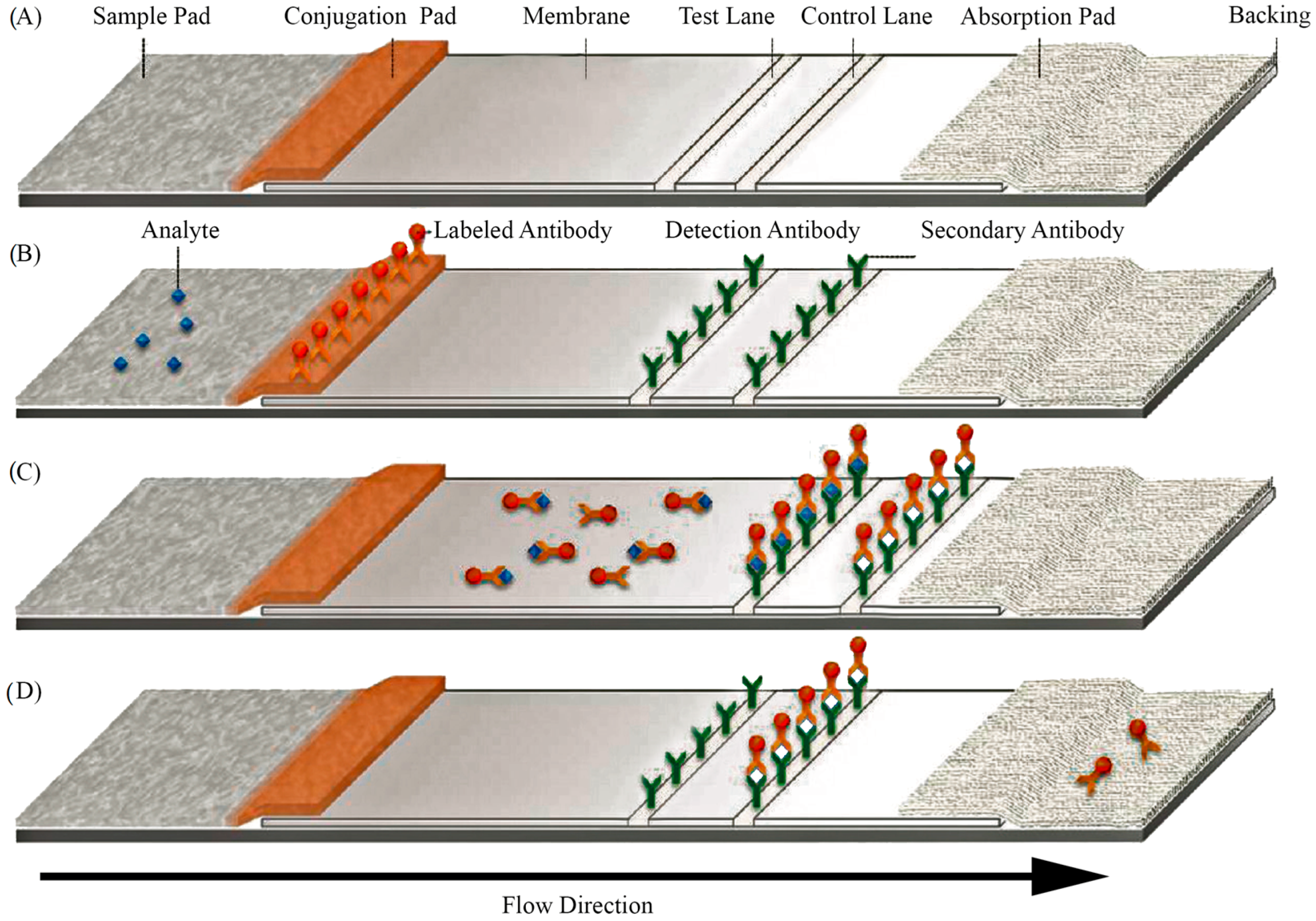
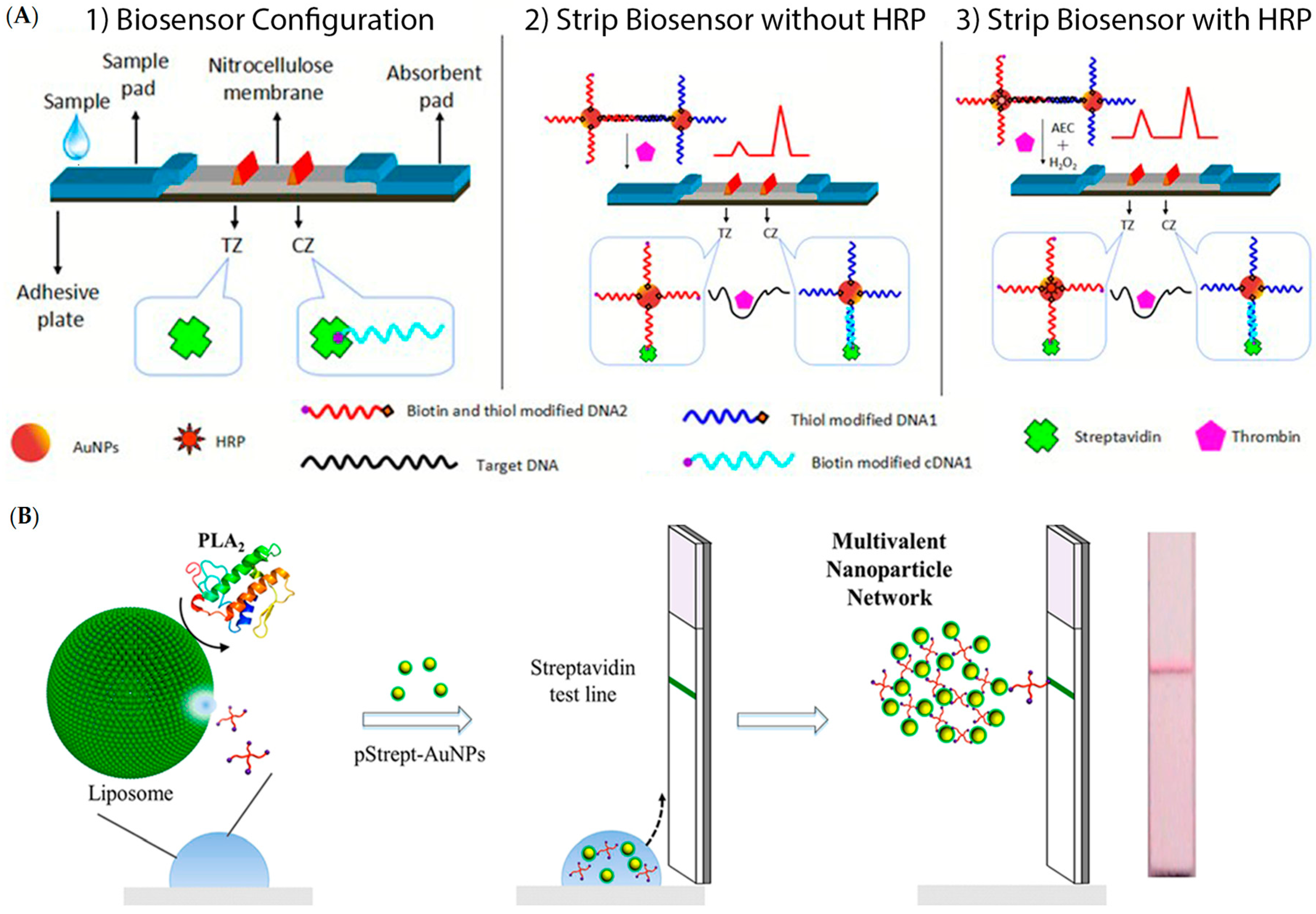
3.1. Lateral Flow Assays (LFAs)
3.2. Microfluidics
3.3. Screen Printed Electrodes
3.4. Smartphone Assisted Readout
4. AuNPs Based Systems Established at POC
Achievements and Challenges to Overcome
5. Conclusions and Outlook
Acknowledgments
Conflicts of Interest
References
- Urdea, M.; Penny, L.A.; Olmsted, S.S.; Giovanni, M.Y.; Kaspar, P.; Shepherd, A.; Wilson, P.; Dahl, C.A.; Buchsbaum, S.; Moeller, G.; et al. Requirements for high impact diagnostics in the developing world. Nature 2006, 444, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Drain, P.K.; Hyle, E.P.; Noubary, F.; Freedberg, K.A.; Wilson, D.; Bishai, W.R.; Rodriguez, W.; Bassett, I.V. Diagnostic point-of-care tests in resource-limited settings. Lancet Infect. Dis. 2014, 14, 239–249. [Google Scholar] [CrossRef]
- Srinivasan, B.; Tung, S. Development and applications of portable biosensors. J. Lab. Autom. 2015, 20, 365–389. [Google Scholar] [CrossRef] [PubMed]
- St John, A.; Price, C. The march of technology through the clinical laboratory and beyond. Clin. Biochem. Rev. 2014, 35, 139–141. [Google Scholar] [PubMed]
- Syedmoradi, L.; Daneshpour, M.; Alvandipour, M.; Gomez, F.A.; Hajghassem, H.; Omidfar, K. Point of care testing: The impact of nanotechnology. Biosens. Bioelectron. 2017, 87, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Pelton, M.; Aizpurua, J.; Bryant, G. Metal-nanoparticle plasmonics. Laser Photonic Rev. 2008, 2, 136–159. [Google Scholar] [CrossRef]
- Herizchi, R.; Abbasi, E.; Milani, M.; Akbarzadeh, A. Current methods for synthesis of gold nanoparticles. Artif. Cells Nanomed. Biotechnol. 2016, 44, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Eustis, S.; El-Sayed, M.A. Why gold nanoparticles are more precious than pretty gold: Noble metal surface plasmon resonance and its enhancement of the radiative and nonradiative properties of nanocrystals of different shapes. Chem. Soc. Rev. 2006, 35, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Link, S.; El-Sayed, M.A. Optical properties and ultrafast dynamics of metallic nanocrystals. Annu. Rev. Phys. Chem. 2003, 54, 331–366. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jia, Q.; Yang, C.; Qiao, R.; Jing, L.; Wang, L.; Xu, C.; Gao, M. Lateral flow immunochromatographic assay for sensitive pesticide detection by using Fe3O4 nanoparticle aggregates as color reagents. Anal. Chem. 2011, 83, 6778–6784. [Google Scholar] [CrossRef] [PubMed]
- Xianyu, Y.; Wang, Z.; Jiang, X. A plasmonic nanosensor for immunoassay via enzyme-triggered click chemistry. ACS Nano 2014, 8, 12741–12747. [Google Scholar] [CrossRef] [PubMed]
- Lequin, R.M. Enzyme immunoassay (EIA): Enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef] [PubMed]
- De la Rica, R.; Stevens, M.M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat. Nanotechnol. 2012, 7, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, J.; Song, S.; Li, D.; Fan, C. Biomolecular sensing via coupling DNA-based recognition with gold nanoparticles. J. Phys. D Appl. Phys. 2009, 42, 203001. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer Science & Business Media: Berlin, Germany, 2013; pp. 5–7. [Google Scholar]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2014 update on diagnosis, monitoring, and management. Am. J. Hematol. 2014, 89, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Boyd, S.D. Diagnostic applications of high-throughput DNA sequencing. Annu. Rev. Pathol. 2013, 8, 381–410. [Google Scholar] [CrossRef] [PubMed]
- Tsagarakis, N.J.; Kentrou, N.A.; Kakiopoulos, G.; Androutsos, G.; Galanopoulos, A.; Michaelidis, C.; Rontogianni, D.; Tolis, A.; Chini, S.; Gortzolidis, G.; et al. Flow cytometry as a diagnostic tool in the early diagnosis of aggressive lymphomas mimicking life-threatening infection. Case Rep. Med. 2011, 2011, 743817. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.; Myneedu, V.P. Microscopy as a diagnostic tool in pulmonary tuberculosis. Int. J. Mycobacteriol. 2015, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sieroń, A.; Sieroń-Stołtny, K.; Kawczyk-Krupka, A.; Latos, W.; Kwiatek, S.; Straszak, D.; Bugaj, A.M. The role of fluorescence diagnosis in clinical practice. Onco Targets Ther. 2013, 6, 977–982. [Google Scholar] [PubMed]
- Kang, K.A.; Wang, J.; Jasinski, J.B.; Achilefu, S. Fluorescence manipulation by gold nanoparticles: From complete quenching to extensive enhancement. J. Nanobiotechnol. 2011, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Rosa, J.P.; Lima, J.C.; Baptista, P.V. Experimental photophysical characterization of fluorophores in the vicinity of gold nanoparticles. Nanotechnology 2011, 22, 415202. [Google Scholar] [CrossRef] [PubMed]
- Degliangeli, F.; Kshirsagar, P.; Brunetti, V.; Pompa, P.P.; Fiammengo, R. Absolute and direct microRNA quantification using DNA-gold nanoparticle probes. J. Am. Chem. Soc. 2014, 136, 2264–2267. [Google Scholar] [CrossRef] [PubMed]
- Beni, V.; Zewdu, T.; Joda, H.; Katakis, I.; O’Sullivan, C.K. Gold nanoparticle fluorescent molecular beacon for low-resolution DQ2 gene HLA typing. Anal. Bioanal. Chem. 2012, 402, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Chan, C.; Pang, Y.; Ye, W.; Tian, F.; Lyu, J.; Zhang, Y.; Yang, M. A fluorescence resonance energy transfer (FRET) biosensor based on graphene quantum dots (GQDs) and gold nanoparticles (AuNPs) for the detection of mecA gene sequence of Staphylococcus aureus. Biosens. Bioelectron. 2015, 67, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Chinen, A.B.; Guan, C.M.; Ferrer, J.R.; Barnaby, S.N.; Merkel, T.J.; Mirkin, C.A. Nanoparticle probes for the detection of cancer biomarkers, cells, and tissues by fluorescence. Chem. Rev. 2015, 115, 10530–10574. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Cho, I.H.; Park, J.N.; Seo, S.M.; Paek, S.H. A high-performance fluorescence immunoassay based on the relaxation of quenching, exemplified by detection of cardiac troponin I. Sensors 2016, 16, 669. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Kong, T.; Zhang, D.; Zhang, J.; Cheng, G. Label-Free MicroRNA detection based on fluorescence quenching of gold nanoparticles with a competitive hybridization. Anal. Chem. 2015, 87, 10822–10829. [Google Scholar] [CrossRef] [PubMed]
- Muehlethaler, C.; Leona, M.; Lombardi, J.R. Review of surface enhanced Raman scattering applications in forensic science. Anal. Chem. 2015, 88, 152–169. [Google Scholar] [CrossRef] [PubMed]
- Alonso-González, P.; Albella, P.; Schnell, M.; Chen, J.; Huth, F.; García-Etxarri, A.; Casanova, F.; Golmar, L.; Arzubiaga, L.; Hueso, L.E.; et al. Resolving the electromagnetic mechanism of surface-enhanced light scattering at single hot spots. Nat. Commun. 2012, 3, 684. [Google Scholar] [Green Version]
- Bantz, K.C.; Meyer, A.F.; Wittenberg, N.J.; Im, H.; Kurtuluş, Ö.; Lee, S.H.; Lindquist, N.C.; Oh, S.; Haynes, C.L. Recent progress in SERS biosensing. Phys. Chem. Chem. Phys. 2011, 13, 11551–11567. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Wang, H.N.; Scaffidi, J. Plasmonic nanoprobes for SERS biosensing and bioimaging. J. Biophotonics 2010, 3, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Podstawka, E.; Ozaki, Y.; Proniewicz, L.M. Part III: Surface-enhanced Raman scattering of amino acids and their homodipeptide monolayers deposited onto colloidal gold surface. Appl. Spectrosc. 2005, 59, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Lyandres, O.; Yuen, J.M.; Shah, N.C.; VanDuyne, R.P.; Walsh, J.T., Jr.; Glucksberg, M.R. Progress toward an in vivo surface-enhanced Raman spectroscopy glucose sensor. Diabetes Technol. Ther. 2008, 10, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.Y.C.; Xiao, W.; West, N.P.; Wee, E.J.; Wang, Y.; Trau, M. Rapid, single-cell electrochemical detection of Mycobacterium tuberculosis using colloidal gold nanoparticles. Anal. Chem. 2015, 87, 10613–10618. [Google Scholar] [CrossRef] [PubMed]
- He, J.L.; Tian, Y.F.; Cao, Z.; Zou, W.; Sun, X. An electrochemical immunosensor based on gold nanoparticle tags for picomolar detection of c-Myc oncoprotein. Sens. Actuators B Chem. 2013, 181, 835–841. [Google Scholar] [CrossRef]
- Liu, X.; Wong, D.K. Picogram-detection of estradiol at an electrochemical immunosensor with a gold nanoparticle Protein G-(LC-SPDP)-scaffold. Talanta 2009, 77, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Zamborini, F.P.; Leopold, M.C.; Hicks, J.F.; Kulesza, P.J.; Malik, M.A.; Murray, R.W. Electron hopping conductivity and vapor sensing properties of flexible network polymer films of metal nanoparticles. J. Am. Chem. Soc. 2002, 124, 8958–8964. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.; Agasti, S.S.; Kim, C.; Li, X.; Rotello, V.M. Gold nanoparticles in chemical and biological sensing. Chem. Rev. 2012, 112, 2739–2779. [Google Scholar] [CrossRef] [PubMed]
- Pingarrón, J.M.; Yañez-Sedeño, P.; González-Cortés, A. Gold nanoparticle-based electrochemical biosensors. Electrochim. Acta 2008, 53, 5848–5866. [Google Scholar] [CrossRef]
- Li, Y.; Schluesener, H.J.; Xu, S. Gold nanoparticle-based biosensors. Gold Bull. 2010, 43, 29–41. [Google Scholar] [CrossRef]
- Uludag, Y.; Köktürk, G. Determination of prostate-specific antigen in serum samples using gold nanoparticle based amplification and lab-on-a-chip based amperometric detection. Microchim. Acta 2015, 182, 1685–1691. [Google Scholar] [CrossRef]
- Maltez-da Costa, M.; de la Escosura-Muñiz, A.; Nogués, C.; Barrios, L.; Ibáñez, E.; Merkoçi, A. Simple monitoring of cancer cells using nanoparticles. Nano Lett. 2012, 12, 4164–4171. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, X. Gold nanoparticles for photoacoustic imaging. Nanomedicine 2015, 10, 299–320. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Jung, S.Y.; Lee, S.J. Gold nanoparticle contrast agents in advanced X-ray imaging technologies. Molecules 2013, 108, 5858–5890. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Dias, J.T.; Grazú, V.; Moros, M.; Baptista, P.V.; de la Fuente, J.M. Revisiting 30 years of biofunctionalization and surface chemistry of inorganic nanoparticles for nanomedicine. Front. Chem. 2014, 2, 48. [Google Scholar] [CrossRef] [PubMed]
- Doria, G.; Conde, J.; Veigas, B.; Giestas, L.; Almeida, C.; Assunção, M.; Rosa, J.; Baptista, P.V. Noble metal nanoparticles for biosensing applications. Sensors 2012, 12, 1657–1687. [Google Scholar] [CrossRef] [PubMed]
- Wetmur, J.G.; Fresco, J. DNA probes: Applications of the principles of nucleic acid hybridization. Crit. Rev. Biochem. Mol. Biol. 2008, 26, 227–259. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Ru, K.; Zhang, L.; Huang, Y.; Zhu, X.; Liu, H.; Zetterberg, A.; Cheng, T.; Miao, W. Fluorescence in situ hybridization (FISH): An increasingly demanded tool for biomarker research and personalized medicine. Biomark. Res. 2014, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Garibyan, L.; Avashia, N. Research techniques made simple: Polymerase chain reaction (PCR). J. Investig. Dermatol. 2013, 133, e6. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Dirks, R.M.; Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275–15278. [Google Scholar] [CrossRef] [PubMed]
- Trevino, V.; Falciani, F.; Barrera-Saldaña, H.A. DNA microarrays: A powerful genomic tool for biomedical and clinical research. Mol. Med. 2007, 13, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Arora, A.; Wengel, J.; Maiti, S. Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes. Biochemistry 2006, 45, 7347–7355. [Google Scholar] [CrossRef] [PubMed]
- Braasch, D.A.; Corey, D.R. Locked nucleic acid (LNA): Fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001, 8, 1–7. [Google Scholar] [CrossRef]
- Pande, V.; Nilsson, L. Insights into structure, dynamics and hydration of locked nucleic acid (LNA) strand-based duplexes from molecular dynamics simulations. Nucleic Acids Res. 2008, 36, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Mukhopadhyay, R. Locked Nucleic Acid (LNA)-based Nucleic Acid Sensors. J. Bioanal. Biomed. 2013, 5, e114. [Google Scholar]
- Shakeel, S.; Karim, S.; Ali, A. Peptide nucleic acid (PNA)—A review. J. Chem. Technol. Biotechnol. 2006, 81, 892–899. [Google Scholar] [CrossRef]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Elghanian, R.; Storhoff, J.J.; Mucic, R.C.; Letsinger, R.L.; Mirkin, C.A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 1997, 277, 1078–1081. [Google Scholar] [CrossRef] [PubMed]
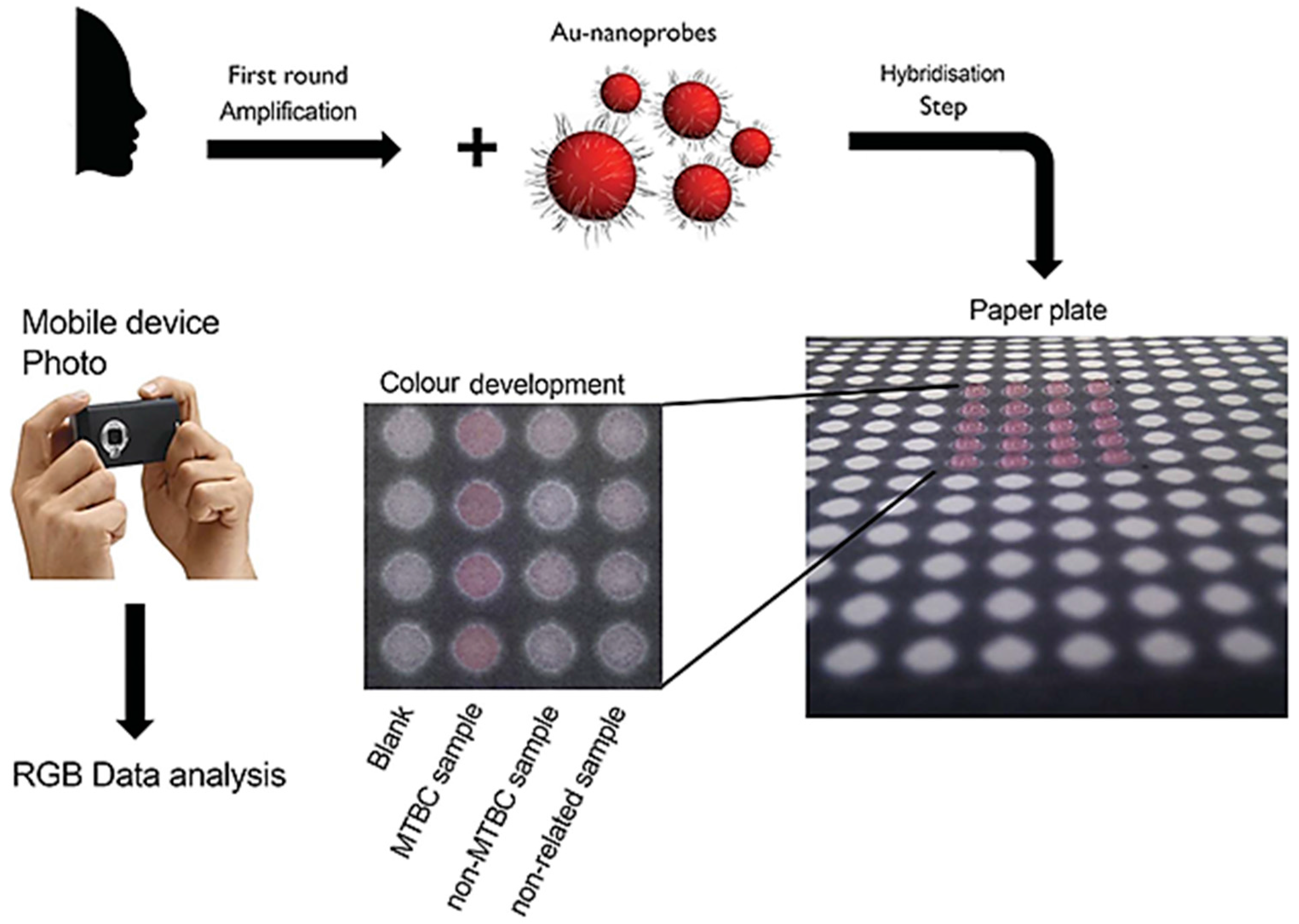
- Veigas, B.; Jacob, J.M.; Costa, M.N.; Santos, D.S.; Viveiros, M.; Inácio, J.; Martins, R.; Barquinha, P.; Fortunato, E.; Baptista, P.V. Gold on paper-paper platform for Au-nanoprobe TB detection. Lab Chip 2012, 12, 4802–4808. [Google Scholar] [CrossRef] [PubMed]
- Vinhas, R.; Correia, C.; Ribeiro, P.; Lourenço, A.; Botelho de Sousa, A.; Fernandes, A.R.; Baptista, P.V. Colorimetric assessment of BCR-ABL1 transcripts in clinical samples via gold nanoprobes. Anal. Bioanal. Chem. 2016, 408, 5277–5284. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xu, Y.; Ferhan, A.R.; Chen, G.; Kim, D.H. Oriented gold nanoparticle aggregation for colorimetric sensors with surprisingly high analytical figures of merit. J. Am. Chem. Soc. 2013, 135, 12338–12345. [Google Scholar] [CrossRef] [PubMed]
- Kato, D.; Oishi, M. Ultrasensitive detection of DNA and RNA based on enzyme-free click chemical ligation chain reaction on dispersed gold nanoparticles. ACS Nano 2014, 8, 9988–9997. [Google Scholar] [CrossRef] [PubMed]
- Liandris, E.; Gazouli, M.; Andreadou, M.; Čomor, M.; Abazovic, N.; Sechi, L.A.; Ikonomopoulos, J. Direct detection of unamplified DNA from pathogenic mycobacteria using DNA-derivatized gold nanoparticles. J. Microbiol. Methods 2009, 78, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Rozenblum, G.T.; Lopez, V.G.; Vitullo, A.D.; Radrizzani, M. Aptamers: Current challenges and future prospects. Expert Opin. Drug Discov. 2016, 11, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, M.; Ellington, A.D. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal. Bioanal. Chem. 2008, 390, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Mao, X.; Zeng, Q.; Wang, S.; Kawde, A.N.; Liu, G. Aptamer-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for protein analysis. Anal. Chem. 2009, 81, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Ahn, J.Y.; Lee, S.H.; Sekhon, S.S.; Kim, D.G.; Min, J.; Kim, Y.H. Aptamer-based sandwich assay and its clinical outlooks for detecting lipocalin-2 in hepatocellular carcinoma (HCC). Sci. Rep. 2015, 5, 10897. [Google Scholar] [CrossRef] [PubMed]
- Lavu, P.S.R.; Mondal, B.; Ramlal, S.; Murali, H.S.; Batra, H.V. Selection and characterization of aptamers using a modified whole cell bacterium SELEX for the detection of Salmonella enterica serovar typhimurium. ACS. Comb. Sci. 2016, 18, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zu, Y. A highlight of recent advances in aptamer technology and its application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Tighe, P.J.; Ryder, R.R.; Todd, I.; Fairclough, L.C. ELISA in the multiplex era: Potentials and pitfalls. Proteom. Clin. Appl. 2015, 9, 406–422. [Google Scholar] [CrossRef] [PubMed]
- Chard, T. Pregnancy tests: A review. Hum. Reprod. 1992, 7, 701–710. [Google Scholar] [PubMed]
- Parolo, C.; de la Escosura-Muñiz, A.; Merkoçi, A. Enhanced lateral flow immunoassay using gold nanoparticles loaded with enzymes. Biosens. Bioelectron. 2013, 40, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Y.Q.; Ye, B.C. Colorimetric assay for parallel detection of Cd2+, Ni2+ and Co2+ using peptide-modified gold nanoparticles. Analyst 2012, 137, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Li, X.; Liu, X.; Wang, J.; Wang, Z. Designing bifunctionalized gold nanoparticle for colorimetric detection of Pb2+ under physiological condition. Biosens. Bioelectron. 2012, 31, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lévy, R.; Fernig, D.G.; Brust, M. Kinase-catalyzed modification of gold nanoparticles: A new approach to colorimetric kinase activity screening. J. Am. Chem. Soc. 2006, 128, 2214–2215. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Kawde, A.N.; Daud, M. Designs, formats and applications of lateral flow assay: A literature review. J. Saudi Chem. Soc. 2015, 19, 689–705. [Google Scholar] [CrossRef]
- Sun, J.; Xianyu, Y.; Jiang, X. Point-of-care biochemical assays using gold nanoparticle-implemented microfluidics. Chem. Soc. Rev. 2014, 43, 6239–6253. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zapatero-Rodríguez, J.; Estrela, P.; O’Kennedy, R. Point-of-care diagnostics in low resource settings: Present status and future role of microfluidics. Biosensors 2015, 5, 577–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno) assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Rivas, L.; Medina-Sánchez, M.; de la Escosura-Muñiz, A.; Merkoçi, A. Improving sensitivity of gold nanoparticle-based lateral flow assays by using wax-printed pillars as delay barriers of microfluidics. Lab Chip 2014, 14, 4406–4414. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, B. Lateral flow immunoassay systems: Evolution from the current state of the art to the next generation of highly sensitive, quantitative rapid assays. In The Immunoassay Handbook: Theory and Applications of Ligand Binding, ELISA and Related Techniques, 4th ed.; Wild, D., Ed.; Newnes: Boston, MA, USA, 2013; pp. 89–107. [Google Scholar]
- Hu, J.; Wang, L.; Li, F.; Han, Y.L.; Lin, M.; Lu, T.J.; Xu, F. Oligonucleotide-linked gold nanoparticle aggregates for enhanced sensitivity in lateral flow assays. Lab Chip 2013, 13, 4352–4357. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Du, T.E.; Meng, L.; Song, T. Novel gold nanoparticle trimer reporter probe combined with dry-reagent cotton thread immunoassay device for rapid human ferritin test. Anal. Chim. Acta 2015, 889, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Wang, J.; Tang, Z.; Pounds, J.G.; Lin, Y. Rapid and sensitive detection of protein biomarker using a portable fluorescence biosensor based on quantum dots and a lateral flow test strip. Anal. Chem. 2010, 82, 7008–7014. [Google Scholar] [CrossRef] [PubMed]
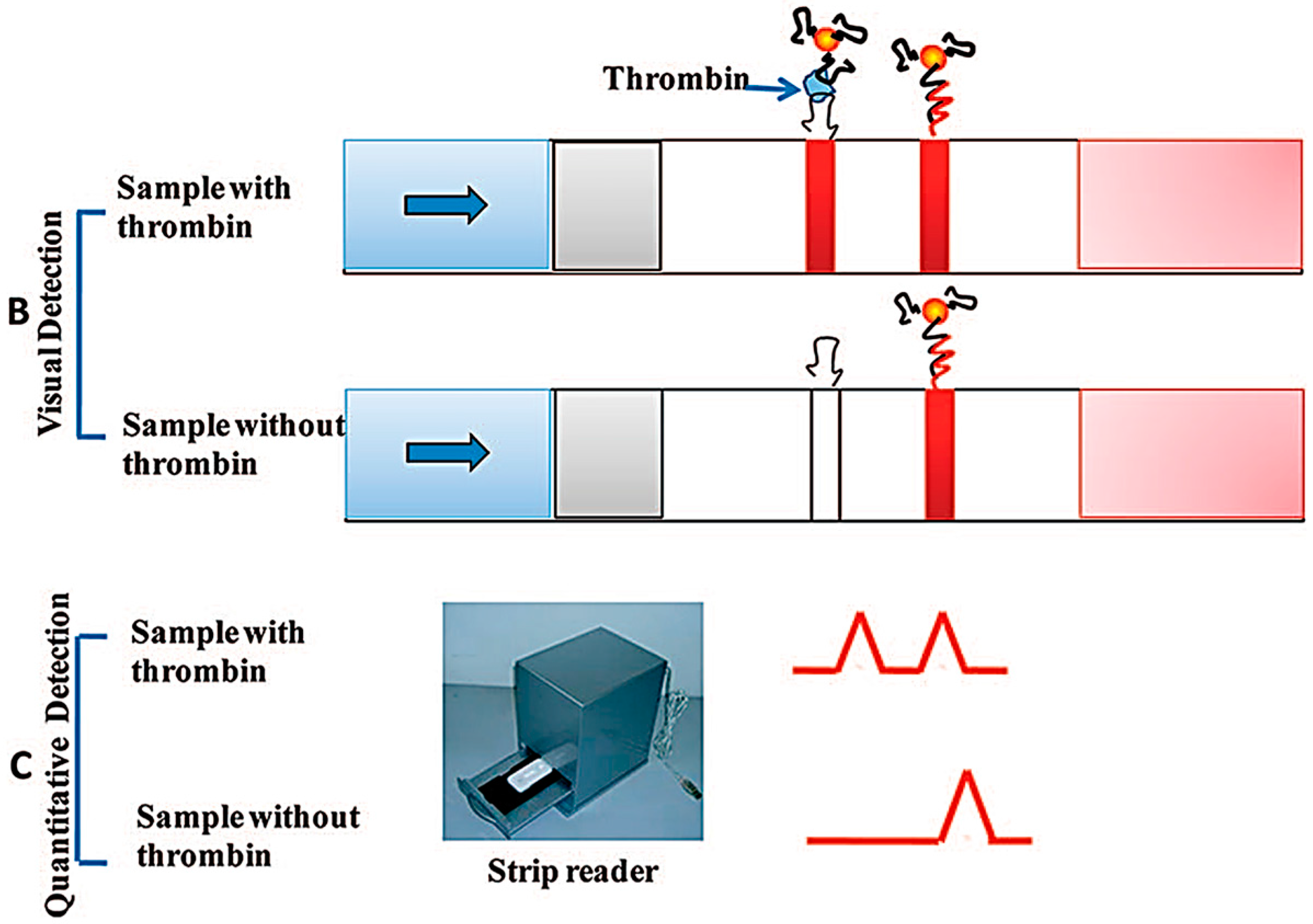
- Qin, C.; Wen, W.; Zhang, X.H.; Gu, H.S.; Wang, S.F. Visual detection of thrombin using a strip biosensor through aptamer-cleavage reaction with enzyme catalytic amplification. Analyst 2015, 140, 7710–7717. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.; Lin, Y.; Burnapp, M.; Bentham, A.; Hillier, D.; Zabron, A.; Khan, S.; Tyreman, M.; Stevens, M.M. Multivalent nanoparticle networks enable point-of-care detection of human phospholipase-A2 in serum. ACS Nano 2015, 9, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Pastuszczak, M.; Wojas-Pelc, A. Current standards for diagnosis and treatment of syphilis: Selection of some practical issues, based on the European (IUSTI) and US (CDC) guidelines. Postep. Dermatol. Alergol. 2013, 30, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Ma, J.; Zhang, Q.; Li, N.; Yang, J.; Raju, P.A.; Peng, M.; Luo, Y.; Hui, W.; Chen, C.; et al. Polyelectrolyte-coated gold magnetic nanoparticles for immunoassay development: Toward point of care diagnostics for syphilis screening. Anal. Chem. 2013, 85, 6688–6695. [Google Scholar] [CrossRef] [PubMed]
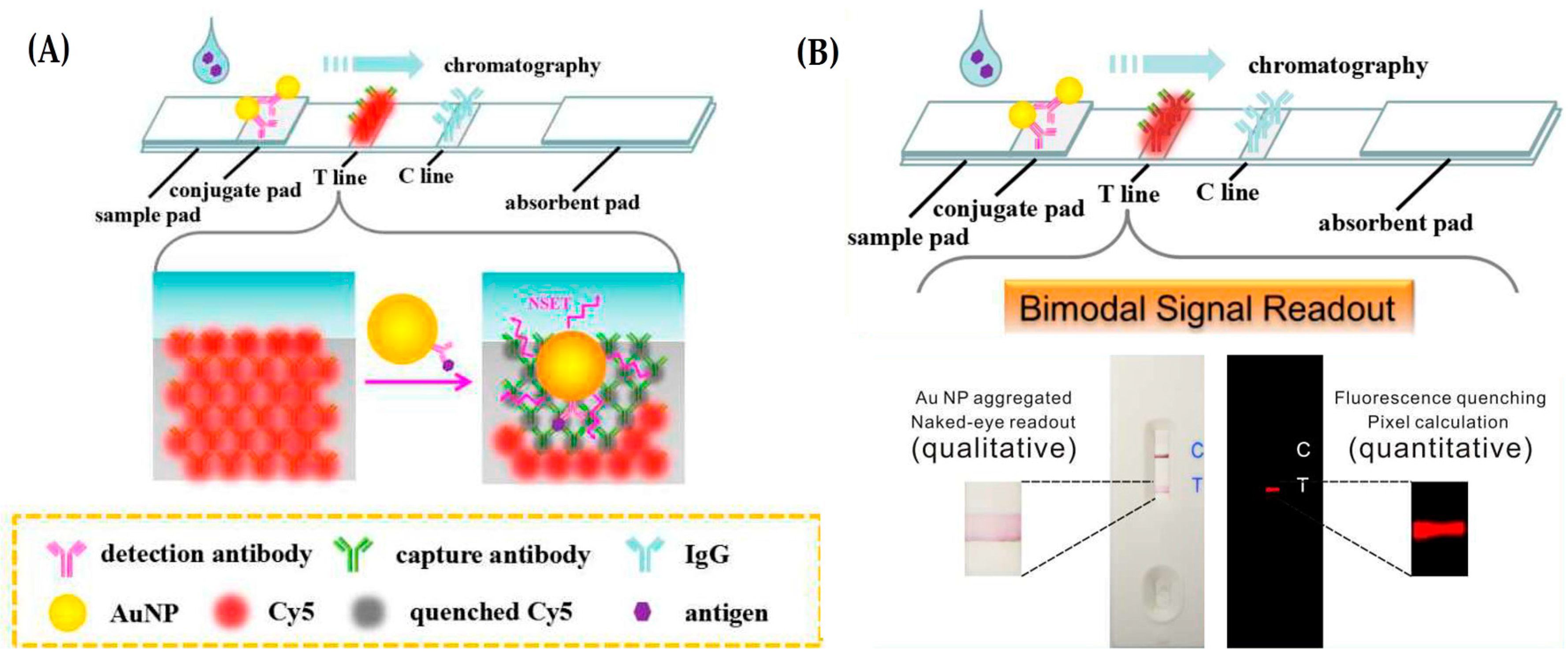
- Yao, Y.; Guo, W.; Zhang, J.; Wu, Y.; Fu, W.; Liu, T.; Wu, X.; Wang, H.; Gong, X.; Liang, X.; et al. Reverse fluorescence enhancement and colorimetric bimodal signal readout immunochromatography test strip for ultrasensitive large-scale screening and postoperative monitoring. ACS Appl. Mater. Interfaces 2016, 8, 22963–22970. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.; Fulton, A.; Beebe, D. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Foudeh, A.; Didar, T.; Veresa, T.; Tabrizian, M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 2012, 12, 3249–3266. [Google Scholar] [CrossRef] [PubMed]
- Sanghavi, B.; Moore, J.; Chávez, J.; Hagen, J.; Kelley-Loughnane, N.; Chou, C.; Swami, N. Aptamer-functionalized nanoparticles for surface immobilization-free electrochemical detection of cortisol in a microfluidic device. Biosens. Bioelectron. 2016, 78, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Torul, H.; Çiftçi, H.; Çetin, D.; Suludere, Z.; Boyacı, I.; Tamer, U. Paper membrane-based SERS platform for the determination of glucose in blood samples. Anal. Bioanal. Chem. 2015, 407, 8243–8251. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Huang, Y.; Ma, Y.; Jia, S.; Gao, M.; Li, J.; Zhang, H.; Xu, D.; Wu, M.; Chen, Y.; et al. Design and synthesis of target-responsive aptamer-cross-linked hydrogel for visual quantitative detection of Ochratoxin A. ACS Appl. Mater. Interface 2015, 7, 6982–6990. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.; Dong, T. An integrated passive-flow microfluidic biosensor with organic photodiodes for ultra-sensitive pathogen detection in water. IEEE Eng. Med. Biol. Soc. Conf. Proc. 2014, 2014, 4411–4414. [Google Scholar]
- Ölcer, Z.; Esen, E.; Ersoy, A.; Budak, S.; Kaya, D.S.; Gök, M.Y.; Barut, S.; Üstek, D.; Uludag, Y. Microfluidics and nanoparticles based amperometric biosensor for the detection of cyanobacteria (Planktothrix agardhii NIVA-CYA 116) DNA. Biosens. Bioelectron. 2015, 70, 426–432. [Google Scholar]
- Kara, A.; Rouillard, C.; Mathault, J.; Boisvert, M.; Tessier, F.; Landari, H.; Melki, I.; Laprise-Pelletier, M.; Boisselier, E.; Fortin, M.; et al. Towards a multifunctional electrochemical sensing and niosome generation lab-on-chip platform based on a plug-and-play concept. Sensors 2016, 16, 778. [Google Scholar] [CrossRef] [PubMed]
- Sedighi, A.; Li, P. High-throughput DNA array for SNP detection of KRAS gene using a centrifugal microfluidic device. Methods Mol. Biol. 2016, 1368, 133–141. [Google Scholar] [PubMed]
- Bernacka-Wojcik, I.; Águas, H.; Carlos, F.F.; Lopes, P.; Wojcik, P.J.; Costa, M.N.; Veigas, B.; Igreja, R.; Fortunato, E.; Baptista, P.V.; et al. Single nucleotide polymorphism detection using gold nanoprobes and bio-microfluidic platform with embedded microlenses. Biotechnol. Bioeng. 2015, 112, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Jana, N. Paper-based microfluidic approach for surface-enhanced Raman spectroscopy and highly reproducible detection of proteins beyond picomolar concentration. ACS Appl. Mater. Interfaces 2015, 7, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.; Sharma, M.; Pandey, P.; Sekhar, K. Comparison of different carbon ink based screen-printed electrodes towards amperometric immunosensing. World J. Microbiol. Biotechnol. 2006, 22, 1135–1143. [Google Scholar] [CrossRef]
- Bain, C.; Troughton, E.; Tao, Y.; Evall, J.; Whitesides, G.; Nuzzo, R. Formation of monolayer films by the spontaneous assembly of organic thiols from solution onto gold. J. Am. Chem. Soc. 1989, 111, 321–335. [Google Scholar] [CrossRef]
- Taleat, Z.; Khoshroo, A.; Mazloum-Ardakani, M. Screen-printed electrodes for biosensing: A review (2008–2013). Microchim. Acta 2014, 181, 865–891. [Google Scholar] [CrossRef]
- Ahmed, M.U.; Hossain, M.M.; Safavieh, M.; Wong, Y.L.; Rahman, I.A.; Zourob, M.; Tamiya, E. Toward the development of smart and low cost point-of-care biosensors based on screen printed electrodes. Crit. Rev. Biotechnol. 2016, 36, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Khorsand, F.; Azizi, M.; Naeemy, A.; Larijani, B.; Omidfar, K. An electrochemical biosensor for 3-hydroxybutyrate detection based on screen-printed electrode modified by coenzyme functionalized carbon nanotubes. Mol. Biol. Rep. 2013, 40, 2327–2334. [Google Scholar] [CrossRef] [PubMed]
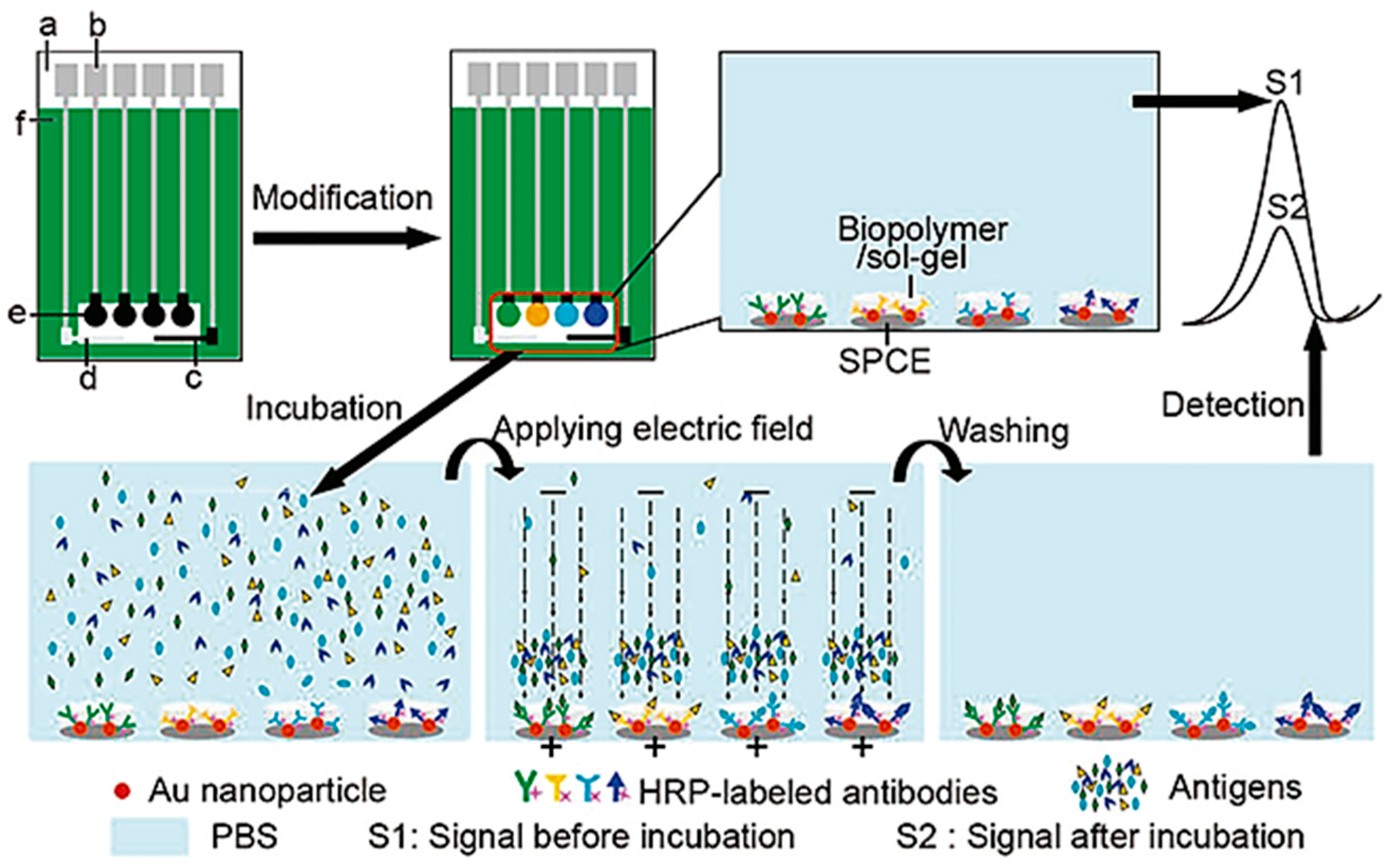
- Wu, J.; Yan, F.; Zhang, X.; Yan, Y.; Tang, J.; Ju, H. Disposable reagentless electrochemical immunosensor array based on a biopolymer/sol-gel membrane for simultaneous measurement of several tumor markers. Clin. Chem. 2008, 54, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Duangkaew, P.; Tapaneeyakorn, S.; Apiwat, C.; Dharakul, T.; Laiwejpithaya, S.; Kanatharana, P.; Laocharoensuk, R. Ultrasensitive electrochemical immunosensor based on dual signal amplification process for p16INK4a cervical cancer detection in clinical samples. Biosens. Bioelectron. 2015, 74, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Electrochemical glucose biosensors. Chem. Rev. 2008, 108, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; O’Dell, D.; Hohenstein, J.; Colt, S.; Mehta, S.; Erickson, D. NutriPhone: A mobile platform for low-cost point-of-care quantification of vitamin B12 concentrations. Sci. Rep. 2016, 6, 28237. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Akay, A.; Wei, H.; Wang, S.; Pingguan-Murphy, B.; Erlandsson, B.E.; Li, X.; Lee, W.; Hu, J.; Wang, L.; et al. Advances in smartphone-based point-of-care diagnostics. Proc. IEEE 2015, 103, 236–247. [Google Scholar] [CrossRef]
- Laksanasopin, T.; Guo, T.W.; Nayak, S.; Sridhara, A.; Xie, S.; Olowookere, O.; Cadinu, P.; Meng, F.; Chee, N.H.; Kim, J.; et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 2015, 7, 273re1. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Chen, P.; Tran, N.T.; Zhang, J.; Chia, W.S.; Boujday, S.; Liedberg, B. Smartphone spectrometer for colorimetric biosensing. Analyst 2012, 141, 3233–3238. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Shi, Z.; Fang, C.; Zhang, Y.; Liu, Y.; Li, C. Disposable lateral flow-through strip for smartphone-camera to quantitatively detect alkaline phosphatase activity in milk. Biosens. Bioelectron. 2015, 69, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Cao, F.; Zheng, W.; Tian, Y.; Xianyu, Y.; Xu, P.; Zhang, W.; Wang, Z.; Deng, K.; Jiang, X. Detection of the nanomolar level of total Cr ((III) and (VI)) by functionalized gold nanoparticles and a smartphone with the assistance of theoretical calculation models. Nanoscale 2015, 7, 2042–2049. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.H.; Chen, W.Y.; Yen, Y.C.; Wang, C.W.; Chang, H.T.; Chen, C.F. Detection of mercury (II) ions using colorimetric gold nanoparticles on paper-based analytical devices. Anal. Chem. 2014, 86, 6843–6849. [Google Scholar] [CrossRef] [PubMed]
- Gulka, C.P.; Swartz, J.D.; Wright, D.W. Ni(II)NTA AuNPs as a low-resource malarial diagnostic platform for the rapid colorimetric detection of Plasmodium falciparum Histidine-Rich Protein-2. Talanta 2015, 135, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Oncescu, V.; Mancuso, M.; Mehta, S.; Erickson, D. A smartphone platform for the quantification of vitamin D levels. Lab Chip 2014, 14, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- Franco, R.; Pedrosa, P.; Carlos, F.F.; Veigas, B.; Baptista, P.V. Gold nanoparticles for DNA/RNA-based diagnostics. In Handbook of Nanoparticles; Aliofkhazraei, M., Ed.; Springer: Berlin, Germany, 2015; pp. 1339–1370. [Google Scholar]
- Baptista, P.; Pereira, E.; Eaton, P.; Doria, G.; Miranda, A.; Gomes, I.; Quaresma, P.; Franco, R. Gold nanoparticles for the development of clinical diagnosis methods. Anal. Bioanal. Chem. 2008, 391, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.K.; Luppa, P.B.; Yeo, L.Y.; Ozcan, A.; Luong, J.H. Emerging technologies for next-generation point-of-care testing. Trends Biotechnol. 2015, 33, 692–705. [Google Scholar] [CrossRef] [PubMed]
- BCC Research Healthcare Report 2014; Technical Report Point of Care Diagnostics—Report Overview; Report Code: HLC043D; BCC Research: Wellesley, MA, USA, 2014.
- St John, A.; Price, C.P. Existing and emerging technologies for point-of-care testing. Clin. Biochem. Rev. 2014, 353, 155–167. [Google Scholar]
- Chan, C.P.Y.; Mak, W.C.; Cheung, K.Y.; Sin, K.K.; Yu, C.M.; Rainer, T.H.; Renneberg, R. Evidence-based point-of-care diagnostics: Current status and emerging technologies. Annu. Rev. Anal. Chem. 2013, 6, 191–211. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.K.; Gibson, C.M.; Branen, J.R.; Aston, D.E.; Branen, A.L.; Hrdlicka, P.J.A. DNA detection on lateral flow test strips: Enhanced signal sensitivity using LNA-conjugated gold nanoparticles. Chem. Commun. 2012, 48, 7714–7716. [Google Scholar] [CrossRef] [PubMed]
- Delaney, K.P.; Branson, B.M.; Uniyal, A.; Phillips, S.; Candal, D.; Owen, S.M.; Kerndt, P.R. Evaluation of the performance characteristics of 6 rapid HIV antibody tests. Clin. Infect. Dis. 2011, 52, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Bertsch, T.; Chapelle, J.P.; Dempfle, C.E.; Giannitsis, E.; Schwab, M.; Zerback, R. Multicentre analytical evaluation of a new point-of-care system for the determination of cardiac and thromboembolic markers. Clin. Lab. 2010, 56, 37–49. [Google Scholar] [PubMed]
- Gils, C.; Ramanathan, R.; Breindahl, T.; Brokner, M.; Christiansen, A.L.; Eng, O.; Hammer, I.; Herrera, C.B.; Jansen, A.; Langsjøen, E.C.; et al. NT-proBNP on Cobas h 232 in point-of-care testing: Performance in the primary health care versus in the hospital laboratory. Scand. J. Clin. Lab. Investig. 2015, 75, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Lefferts, J.A.; Jannetto, P.; Tsongalis, G.J. Evaluation of the Nanosphere Verigene® System System and the Verigene® System F5/F2/MTHFR nucleic acid tests. Exp. Mol. Pathol. 2009, 87, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Maurice, C.B.; Barua, P.K.; Simses, D.; Smith, P.; Howe, J.G.; Stack, G. Comparison of assay systems for warfarin-related CYP2C9 and VKORC1 genotyping. Clin. Chim. Acta 2010, 411, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Malloy, T.F. Nanotechnology regulation: A study in claims making. ACS Nano 2011, 5, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Banoo, S.; Bell, D.; Bossuyt, P.; Herring, A.; Mabey, D.; Poole, F.; Smith, P.G.; Sriram, N.; Wongsrichanalai, C.; Linke, R.; et al. Evaluation of diagnostic tests for infectious diseases: General principles. Nat. Rev. Microbiol. 2008, 8, S16–S28. [Google Scholar] [CrossRef] [PubMed]
| Company | Product Name 1 | Principle of Detection | Sensitivity and Specificity 2 | Results Time |
|---|---|---|---|---|
| Alere, Inc. (Waltham, MA, USA) | Clearview® HIV 1/2 STAT-PAK | Gold-labeled lateral-flow immunoassay | 99.7%/99.9% | 10–15 min |
| Clearview® COMPLETE HIV 1/2 | 99.7%/99.9% | 15 min | ||
| OraSure Technologies, Inc. (Bethlehem, PA, USA) | OraQuick ADVANCE® HIV-1/2 | 98.7%/99.9% | 20–40 min | |
| MedMira, Inc. (Halifax, Nova Scotia, Canada) | Reveal® G3 HIV-1 | 99.8%/99.1% | <3 min | |
| Trinity Biotech Plc (Bray, Republic of Ireland) | Uni-Gold Recombigen™ HIV-1 | 100%/99.8% | 10 min |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cordeiro, M.; Ferreira Carlos, F.; Pedrosa, P.; Lopez, A.; Baptista, P.V. Gold Nanoparticles for Diagnostics: Advances towards Points of Care. Diagnostics 2016, 6, 43. https://doi.org/10.3390/diagnostics6040043
Cordeiro M, Ferreira Carlos F, Pedrosa P, Lopez A, Baptista PV. Gold Nanoparticles for Diagnostics: Advances towards Points of Care. Diagnostics. 2016; 6(4):43. https://doi.org/10.3390/diagnostics6040043
Chicago/Turabian StyleCordeiro, Mílton, Fábio Ferreira Carlos, Pedro Pedrosa, António Lopez, and Pedro Viana Baptista. 2016. "Gold Nanoparticles for Diagnostics: Advances towards Points of Care" Diagnostics 6, no. 4: 43. https://doi.org/10.3390/diagnostics6040043






