Comparison of Direct Sequencing, Real-Time PCR-High Resolution Melt (PCR-HRM) and PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) Analysis for Genotyping of Common Thiopurine Intolerant Variant Alleles NUDT15 c.415C>T and TPMT c.719A>G (TPMT*3C)
Abstract
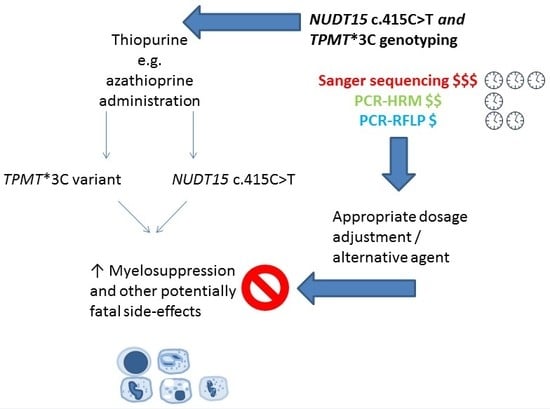
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples and Human Ethics
2.2. DNA Extraction and Sequencing Genotyping
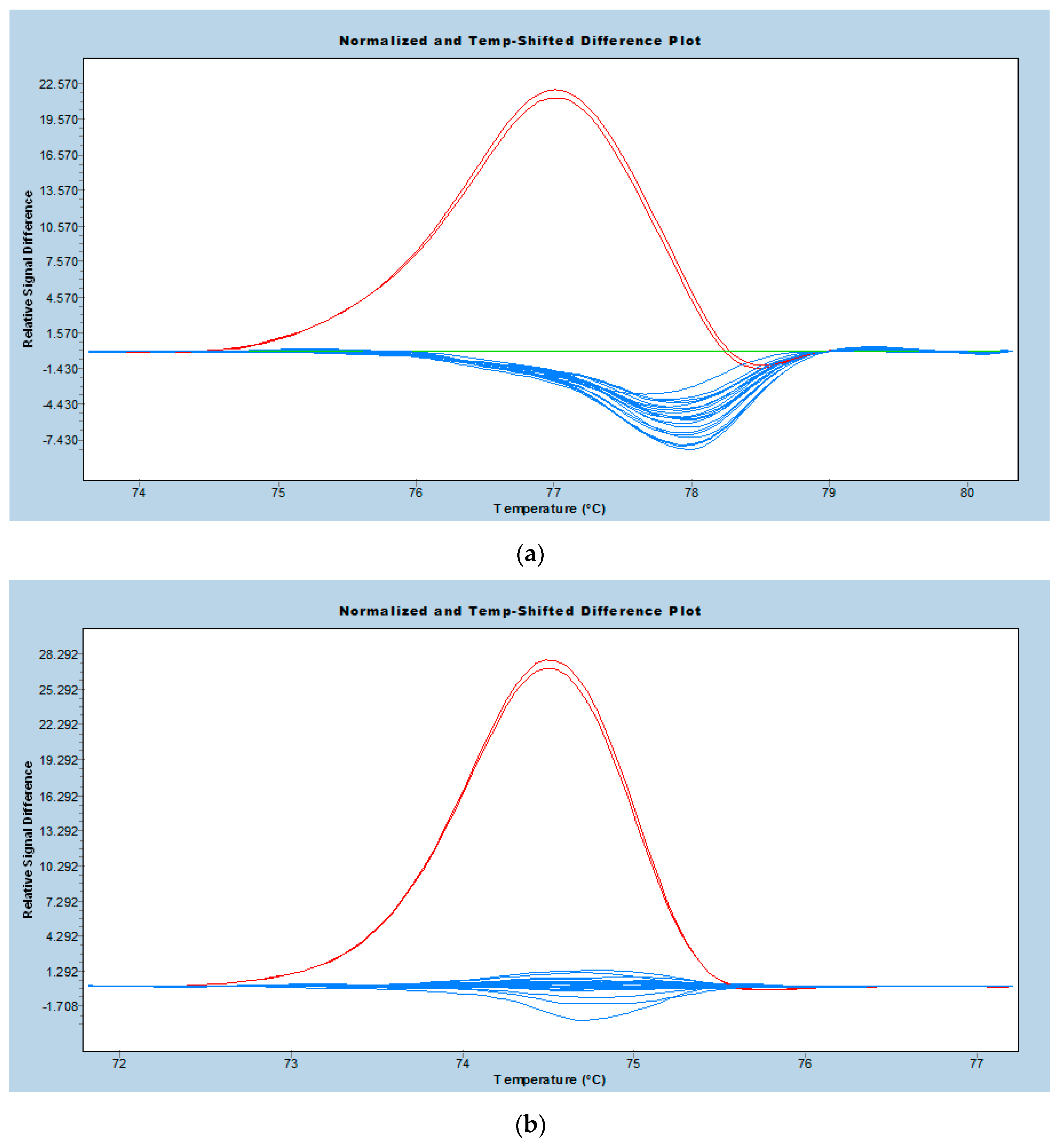
2.3. Real-Time PCR-HRM Analysis
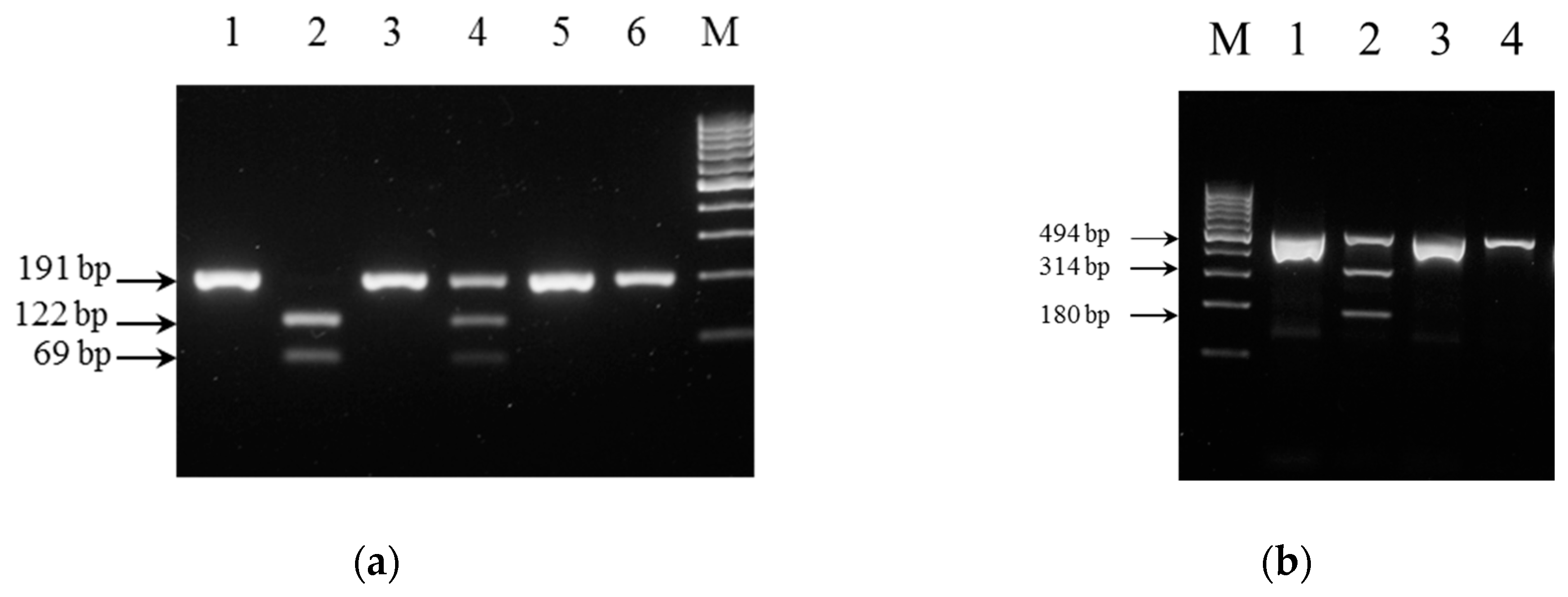
2.4. PCR-RFLP Analysis
3. Results
3.1. Allele Frequency of Variants and Genetic Prevalence of Thiopurine Intolerance
3.2. Performance of Real-Time PCR-HRM Genotyping
3.3. Performance of PCR-RFLP Genotyping
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| TPMT | Thiopurine S-methyltransferase |
| NUDT15 | Nudix hydrolase 15 |
References
- Karran, P.; Attard, N. Thiopurines in current medical practice: Molecular mechanisms and contributions to therapy-related cancer. Nat. Rev. Cancer 2008, 8, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, T.; Nishii, R.; Perez-Andreu, V.; Yang, W.; Klussmann, F.A.; Zhao, X.; Lin, T.-N.; Hoshitsuki, K.; Nersting, J.; Kihira, K.; et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet. 2016, 48, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E. Preventing toxicity with a gene test. Science 2003, 302, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.; Leung, A.; Kwok, J.; Chan, M.; Li, C.; Yuen, Y. NUDT15 variant and thiopurine-induced leukopenia in Hong Kong. Hong Kong Med. J. 2016, 22, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Lennard, L.; Van Loon, J.A.; Weinshilboum, R.M. Pharmacogenetics of acute azathioprine toxicity: Relationship to thiopurine methyltransferase genetic polymorphism. Clin. Pharmacol. Ther. 1989, 46, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.V.; Gardner, E.E.; Sandborn, W.J.; Schmiegelow, K.; Pui, C.-H.; Yee, S.W.; Stein, C.M.; Carrillo, M.; Evans, W.E.; Klein, T.E. Clinical Pharmacogenetics Implementation Consortium Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin. Pharmacol. Ther. 2011, 89, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Krynetski, E.Y.; Tai, H.L.; Yates, C.R.; Fessing, M.Y.; Loennechen, T.; Schuetz, J.D.; Relling, M.V.; Evans, W.E. Genetic polymorphism of thiopurine S-methyltransferase: Clinical importance and molecular mechanisms. Pharmacogenetics 1996, 6, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.V.; Gardner, E.E.; Sandborn, W.J.; Schmiegelow, K.; Pui, C.-H.; Yee, S.W.; Stein, C.M.; Carrillo, M.; Evans, W.E.; Hicks, J.K.; et al. Clinical Pharmacogenetics Implementation Consortium Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin. Pharmacol. Ther. 2013, 93, 324–325. [Google Scholar] [CrossRef] [PubMed]
- Otterness, D.; Szumlanski, C.; Lennard, L.; Klemetsdal, B.; Aarbakke, J.; Park-Hah, J.O.; Iven, H.; Schmiegelow, K.; Branum, E.; O’Brien, J.; et al. Human thiopurine methyltransferase pharmacogenetics: Gene sequence polymorphisms. Clin. Pharmacol. Ther. 1997, 62, 60–73. [Google Scholar] [CrossRef]
- Lennard, L. TPMT in the treatment of Crohn’s disease with azathioprine. Gut 2002, 51, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Takatsu, N.; Matsui, T.; Murakami, Y.; Ishihara, H.; Hisabe, T.; Nagahama, T.; Maki, S.; Beppu, T.; Takaki, Y.; Hirai, F.; et al. Adverse reactions to azathioprine cannot be predicted by thiopurine S-methyltransferase genotype in Japanese patients with inflammatory bowel disease. J. Gastroenterol. Hepatol. 2009, 24, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Tai, H.L.; Krynetski, E.Y.; Yates, C.R.; Loennechen, T.; Fessing, M.Y.; Krynetskaia, N.F.; Evans, W.E. Thiopurine S-methyltransferase deficiency: Two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am. J. Hum. Genet. 1996, 58, 694–702. [Google Scholar] [PubMed]
- Isaza, C.; Henao, J.; López, A.M.; Cacabelos, R. Allelic variants of the thiopurine methyltransferase (TPMT) gene in the Colombian population. Methods Find. Exp. Clin. Pharmacol. 2003, 25, 423–429. [Google Scholar] [CrossRef] [PubMed]
- McLeod, H.L.; Pritchard, S.C.; Githang’a, J.; Indalo, A.; Ameyaw, M.M.; Powrie, R.H.; Booth, L.; Collie-Duguid, E.S. Ethnic differences in thiopurine methyltransferase pharmacogenetics: Evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics 1999, 9, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Hon, Y.Y.; Fessing, M.Y.; Pui, C.H.; Relling, M.V.; Krynetski, E.Y.; Evans, W.E. Polymorphism of the thiopurine S-methyltransferase gene in African-Americans. Hum. Mol. Genet. 1999, 8, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-K.; Hong, M.; Baek, J.; Choi, H.; Zhao, W.; Jung, Y.; Haritunians, T.; Ye, B.D.; Kim, K.-J.; Park, S.H.; et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat. Genet. 2014, 46, 1017–1020. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-T.; Choi, R.; Won, H.-H.; Choe, Y.H.; Kang, B.; Lee, K.; Koo, H.H.; Yoo, K.H.; Kim, Y.-H.; Lee, S.-Y. NUDT15 genotype distributions in the Korean population. Pharmacogenet. Genom. 2017, 27, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Census and Statistics Department. Hong Kong 2011 Population Census—Main Report: Volume I; The HKSAR Government: Hong Kong, China.
- Payne, K.; Newman, W.; Fargher, E.; Tricker, K.; Bruce, I.N.; Ollier, W.E.R. TPMT testing in rheumatology: Any better than routine monitoring? Rheumatol. Oxf. Engl. 2007, 46, 727–729. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, H.W.; Lee, W.; Chun, S.; Min, W.-K. Validation of new allele-specific real-time PCR system for thiopurine methyltransferase genotyping in Korean population. BioMed Res. Int. 2013, 2013, 305704. [Google Scholar] [CrossRef] [PubMed]
- Burchard, P.R.; Abou Tayoun, A.N.; Lefferts, J.A.; Lewis, L.D.; Tsongalis, G.J.; Cervinski, M.A. Development of a rapid clinical TPMT genotyping assay. Clin. Biochem. 2014, 47, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Román, M.; Cabaleiro, T.; Ochoa, D.; Novalbos, J.; Chaparro, M.; Gisbert, J.P.; Abad-Santos, F. Validation of a genotyping method for analysis of TPMT polymorphisms. Clin. Ther. 2012, 34, 878–884. [Google Scholar] [CrossRef] [PubMed]
- Osaki, R.; Imaeda, H.; Ban, H.; Aomatsu, T.; Bamba, S.; Tsujikawa, T.; Sasaki, M.; Fujiyama, Y.; Andoh, A. Accuracy of genotyping using the TaqMan PCR assay for single nucleotide polymorphisms responsible for thiopurine sensitivity in Japanese patients with inflammatory bowel disease. Exp. Ther. Med. 2011, 2, 783–786. [Google Scholar] [PubMed]
- Van Egmond, R.; Barclay, M.L.; Chin, P.K.L.; Sies, C.W.; Florkowski, C.M. Preanalytical stringency: What factors may confound interpretation of thiopurine S-methyl transferase enzyme activity? Ann. Clin. Biochem. 2013, 50, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-C.; Tai, S.-M.; Lee, E.C.-N.; Mak, T.S.-H.; Liu, T.K.-T.; Tang, V.W.-L.; Poon, W.-T. Rapid Identification of Pathogenic Variants in Two Cases of Charcot-Marie-Tooth Disease by Gene-Panel Sequencing. Int. J. Mol. Sci. 2017, 18, 770. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Hwang, E.H.; Park, J.H.; Shin, J.-H.; Kang, B.; Kim, S.-Y. NUDT15 variant is the most common variant associated with thiopurine-induced early leukopenia and alopecia in Korean pediatric patients with Crohn’s disease. Eur. J. Gastroenterol. Hepatol. 2016, 28, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.M.; Bianchi, M.; Guarnieri, C.; Barale, R.; Pacifici, G.M. Genotype-phenotype correlation for thiopurine S-methyltransferase in healthy Italian subjects. Eur. J. Clin. Pharmacol. 2001, 57, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Ewing, B.; Hillier, L.; Wendl, M.C.; Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998, 8, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Liew, M.; Pryor, R.; Palais, R.; Meadows, C.; Erali, M.; Lyon, E.; Wittwer, C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004, 50, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.J.; Landier, W.; Yang, W.; Liu, C.; Hageman, L.; Cheng, C.; Pei, D.; Chen, Y.; Crews, K.R.; Kornegay, N.; et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.M.; Mendes, M.A.S.; Ma, J.D. Thiopurine methyltransferase (TPMT) genotyping to predict myelosuppression risk. PLoS Curr. 2011, 3, RRN1236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guan, Y.; Wu, J.; Xu, A.; Zhou, S.; Huang, M. Phenotyping and genotyping study of thiopurine S-methyltransferase in healthy Chinese children: A comparison of Han and Yao ethnic groups. Br. J. Clin. Pharmacol. 2004, 58, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Vankan, D.M.; Waine, D.R.; Fortes, M.R.S. Real-time PCR genotyping and frequency of the myostatin F94L mutation in beef cattle breeds. Anim. Int. J. Anim. Biosci. 2010, 4, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lo, S.K.; Yuen, K.Y.; Peiris, J.S.; Siau, H.; Chiu, E.K.; Liang, R.H.; Chan, T.K. Detection of CMV DNA in bone marrow transplant recipients: Plasma versus leucocyte polymerase chain reaction. J. Clin. Pathol. 1997, 50, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, F.; Schoendube, J.; Gross, A.; Rath, C.; Niekrawietz, S.; Koltay, P.; Roth, G. Single-cell PCR of genomic DNA enabled by automated single-cell printing for cell isolation. Biosens. Bioelectron. 2015, 69, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Reed, G.H.; Wittwer, C.T. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 2004, 50, 1748–1754. [Google Scholar] [CrossRef] [PubMed]
- Fateh, A.; Aghasadeghi, M.; Siadat, S.D.; Vaziri, F.; Sadeghi, F.; Fateh, R.; Keyvani, H.; Tasbiti, A.H.; Yari, S.; Ataei-Pirkooh, A.; et al. Comparison of Three Different Methods for Detection of IL28 rs12979860 Polymorphisms as a Predictor of Treatment Outcome in Patients with Hepatitis C Virus. Osong Public Health Res. Perspect. 2016, 7, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Garritano, S.; Gemignani, F.; Voegele, C.; Nguyen-Dumont, T.; Le Calvez-Kelm, F.; de Silva, D.; Lesueur, F.; Landi, S.; Tavtigian, S.V. Determining the effectiveness of High Resolution Melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet. 2009, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Krypuy, M.; Newnham, G.M.; Thomas, D.M.; Conron, M.; Dobrovic, A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer 2006, 6, 295. [Google Scholar] [CrossRef] [PubMed]
| Gene Target (dbSNP ID) | Homozygous Wildtype 1 | Heterozygous | Homozygous Mutant | Minor Allele Frequency (95% Confidence Interval) 2 |
|---|---|---|---|---|
| NUDT15 c.415C>T (rs116855232) | 52 (86.7%) | 7 (11.7%) | 1 (1.7%) | 0.075 (0.037–0.14) 3 |
| TPMT c.719A>G (rs1142345) | 59 (98.3%) | 1 (1.7%) | 0 (0.0%) | 0.0083 (0.00044–0.052) 4 |
| Method | Percentage of Mutants Detected 1 | Minimum Amount of Input DNA (ng) 2 | Cost per Sample (USD) 3 | Turn-around Time (h) 4 | Interpretation | Capability to Detect Non-Canonical Mutations |
|---|---|---|---|---|---|---|
| Sanger sequencing | Reference method | ≤1.25 | 16 | 7.5 or 1 working day | Simple, but may be automated and enhanced by software | Yes |
| PCR-high resolution melt (PCR-HRM) | NUDT15 c.415C>T: 100% (8/8) TPMT*3C: 100% (1/1) | 10 | 5 | 2 | Requires special software package provided by realtime PCR platform vendor | Limited 5 |
| PCR-restriction fragment length polymorphism (PCR-RFLP) | NUDT15 c.415C>T: 100% (8/8) TPMT*3C: 100% (1/1) | ≤1.25 | 4 | 3 | Very simple, by comparing band patterns on gel | No |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fong, W.-Y.; Ho, C.-C.; Poon, W.-T. Comparison of Direct Sequencing, Real-Time PCR-High Resolution Melt (PCR-HRM) and PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) Analysis for Genotyping of Common Thiopurine Intolerant Variant Alleles NUDT15 c.415C>T and TPMT c.719A>G (TPMT*3C). Diagnostics 2017, 7, 27. https://doi.org/10.3390/diagnostics7020027
Fong W-Y, Ho C-C, Poon W-T. Comparison of Direct Sequencing, Real-Time PCR-High Resolution Melt (PCR-HRM) and PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) Analysis for Genotyping of Common Thiopurine Intolerant Variant Alleles NUDT15 c.415C>T and TPMT c.719A>G (TPMT*3C). Diagnostics. 2017; 7(2):27. https://doi.org/10.3390/diagnostics7020027
Chicago/Turabian StyleFong, Wai-Ying, Chi-Chun Ho, and Wing-Tat Poon. 2017. "Comparison of Direct Sequencing, Real-Time PCR-High Resolution Melt (PCR-HRM) and PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) Analysis for Genotyping of Common Thiopurine Intolerant Variant Alleles NUDT15 c.415C>T and TPMT c.719A>G (TPMT*3C)" Diagnostics 7, no. 2: 27. https://doi.org/10.3390/diagnostics7020027