New Biomarkers and Diagnostic Tools for the Management of Fever in Low- and Middle-Income Countries: An Overview of the Challenges
Abstract
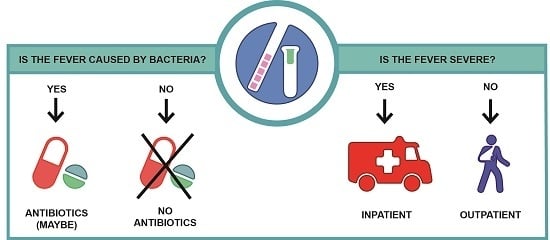
:1. Background
2. Availability of Fever Triage Tests
3. Challenges
3.1. Lack of Reference Tests for Comparative Analysis
3.2. Lack of Regulatory Clarity
3.3. Co-Morbidities in Low- and Middle-Income Countries
3.4. Lack of Compatibility of Clinical Trial Needs with Intended Use Cases
4. Summary
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gething, P.W.; Kirui, V.C.; Alegana, V.A.; Okiro, E.A.; Noor, A.M.; Snow, R.W. Estimating the number of paediatric fevers associated with malaria infection presenting to Africa’s public health sector in 2007. PLoS Med. 2010, 7, e1000301. [Google Scholar] [CrossRef] [PubMed]
- Efstathiou, S.P.; Pefanis, A.V.; Tsiakou, A.G.; Skeva, I.I.; Tsioulos, D.I.; Achimastos, A.D.; Mountokalakis, T.D. Fever of unknown origin: Discrimination between infectious and non-infectious causes. Eur. J. Intern. Med. 2010, 21, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Crump, J.A.; Morrissey, A.B.; Nicholson, W.L.; Massung, R.F.; Stoddard, R.A.; Galloway, R.L.; Ooi, E.E.; Maro, V.P.; Saganda, W.; Kinabo, G.D.; et al. Etiology of severe non-malaria febrile illness in northern Tanzania: A prospective cohort study. PLoS Negl. Trop. Dis. 2013, 7, e2324. [Google Scholar] [CrossRef] [PubMed]
- D’Acremont, V.; Kilowoko, M.; Kyungu, E.; Philipina, S.; Sangu, W.; Kahama-Maro, J.; Lengeler, C.; Cherpillod, P.; Kaiser, L.; Genton, B. Beyond malaria—Causes of fever in outpatient Tanzanian children. N. Engl. J. Med. 2014, 370, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Mayxay, M.; Castonguay-Vanier, J.; Chansamouth, V.; Dubot-Peres, A.; Paris, D.H.; Phetsouvanh, R.; Tangkhabuanbutra, J.; Douangdala, P.; Inthalath, S.; Souvannasing, P.; et al. Causes of non-malarial fever in laos: A prospective study. Lancet Glob. Health 2013, 1, e46–e54. [Google Scholar] [CrossRef]
- Mueller, T.C.; Siv, S.; Khim, N.; Kim, S.; Fleischmann, E.; Ariey, F.; Buchy, P.; Guillard, B.; Gonzalez, I.J.; Christophel, E.M.; et al. Acute undifferentiated febrile illness in rural Cambodia: A 3-year prospective observational study. PLoS ONE 2014, 9, e95868. [Google Scholar] [CrossRef] [PubMed]
- Manock, S.R.; Jacobsen, K.H.; de Bravo, N.B.; Russell, K.L.; Negrete, M.; Olson, J.G.; Sanchez, J.L.; Blair, P.J.; Smalligan, R.D.; Quist, B.K.; et al. Etiology of acute undifferentiated febrile illness in the Amazon basin of Ecuador. Am. J. Trop. Med. Hyg. 2009, 81, 146–151. [Google Scholar] [PubMed]
- Kapasi, A.J.; Dittrich, S.; Gonzalez, I.J.; Rodwell, T.C. Host biomarkers for distinguishing bacterial from non-bacterial causes of acute febrile illness: A comprehensive review. PLoS ONE 2016, 11, e0160278. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, H.; Bruxvoort, K.J.; Cairns, M.E.; Chandler, C.I.; Leurent, B.; Ansah, E.K.; Baiden, F.; Baltzell, K.A.; Bjorkman, A.; Burchett, H.E.; et al. Impact of introduction of rapid diagnostic tests for malaria on antibiotic prescribing: Analysis of observational and randomised studies in public and private healthcare settings. BMJ 2017, 356, j1054. [Google Scholar] [CrossRef] [PubMed]
- WHO. World Malaria Report 2014. Available online: http://www.who.int/malaria/publications/world_malaria_report_2014/wmr-2014-summary-eng.pdf (accessed on 1 June 2017).
- O’Neil, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. 2016. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 1 June 2017).
- Dittrich, S.; Tadesse, B.T.; Moussy, F.; Chua, A.; Zorzet, A.; Tangden, T.; Dolinger, D.L.; Page, A.L.; Crump, J.A.; D’Acremont, V.; et al. Target product profile for a diagnostic assay to differentiate between bacterial and non-bacterial infections and reduce antimicrobial overuse in resource-limited settings: An expert consensus. PLoS ONE 2016, 11, e0161721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The World Bank. Available online: http://data.worldbank.org/income-level/low-and-middle-income (accessed on 30 June 2017).
- Herberg, J.A.; Kaforou, M.; Wright, V.J.; Shailes, H.; Eleftherohorinou, H.; Hoggart, C.J.; Cebey-Lopez, M.; Carter, M.J.; Janes, V.A.; Gormley, S.; et al. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA 2016, 316, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Tsalik, E.L.; Henao, R.; Nichols, M.; Burke, T.; Ko, E.R.; McClain, M.T.; Hudson, L.L.; Mazur, A.; Freeman, D.H.; Veldman, T.; et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 2016, 8, 322ra311. [Google Scholar] [CrossRef] [PubMed]
- Oved, K.; Cohen, A.; Boico, O.; Navon, R.; Friedman, T.; Etshtein, L.; Kriger, O.; Bamberger, E.; Fonar, Y.; Yacobov, R.; et al. A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLoS ONE 2015, 10, e0120012. [Google Scholar] [CrossRef] [PubMed]
- Van Houten, C.B.; de Groot, J.A.; Klein, A.; Srugo, I.; Chistyakov, I.; de Waal, W.; Meijssen, C.B.; Avis, W.; Wolfs, T.F.; Shachor-Meyouhas, Y.; et al. A host-protein based assay to differentiate between bacterial and viral infections in preschool children (opportunity): A double-blind, multicentre, validation study. Lancet Infect. Dis. 2017, 17, 431–440. [Google Scholar] [CrossRef]
- Sweeney, T.E.; Wong, H.R.; Khatri, P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci. Transl. Med. 2016, 8, 346ra391. [Google Scholar] [CrossRef] [PubMed]
- Lubell, Y.; Althaus, T.; Blacksell, S.D.; Paris, D.H.; Mayxay, M.; Pan-Ngum, W.; White, L.J.; Day, N.P.; Newton, P.N. Modelling the impact and cost-effectiveness of biomarker tests as compared with pathogen-specific diagnostics in the management of undifferentiated fever in remote tropical settings. PLoS ONE 2016, 11, e0152420. [Google Scholar] [CrossRef] [PubMed]
- Lubell, Y.; Blacksell, S.D.; Dunachie, S.; Tanganuchitcharnchai, A.; Althaus, T.; Watthanaworawit, W.; Paris, D.H.; Mayxay, M.; Peto, T.J.; Dondorp, A.M.; et al. Performance of c-reactive protein and procalcitonin to distinguish viral from bacterial and malarial causes of fever in Southeast Asia. BMC Infect. Dis. 2015, 15, 511. [Google Scholar] [CrossRef] [PubMed]
- Do, N.T.; Ta, N.T.; Tran, N.T.; Than, H.M.; Vu, B.T.; Hoang, L.B.; van Doorn, H.R.; Vu, D.T.; Cals, J.W.; Chandna, A.; et al. Point-of-care c-reactive protein testing to reduce inappropriate use of antibiotics for non-severe acute respiratory infections in vietnamese primary health care: A randomised controlled trial. Lancet Glob. Health 2016, 4, e633–e641. [Google Scholar] [CrossRef]
- Eden, E.; Srugo, I.; Gottlieb, T.; Navon, R.; Boico, O.; Cohen, A.; Bamberger, E.; Klein, A.; Oved, K. Diagnostic accuracy of a TRAIL, IP-10 and CRP combination for discriminating bacterial and viral etiologies at the Emergency Department. J. Infect. 2016, 73, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Sambursky, R.; Shapiro, N. Evaluation of a combined MXA and CRP point-of-care immunoassay to identify viral and/or bacterial immune response in patients with acute febrile respiratory infection. Eur. Clin. Respir. J. 2015, 2, 28245. [Google Scholar] [CrossRef] [PubMed]
- WHO. Integrated Management of Childhood Illness: Chart Booklet. Available online: http://www.who.int/maternal_child_adolescent/documents/IMCI_chartbooklet/en/ or http://apps.who.int/iris/bitstream/10665/104772/16/9789241506823_Chartbook_eng.pdf (accessed on 7 March 2016).
- WHO. Towards a Grand Convergence for Child Survival and Health. Available online: http://www.who.int/maternal_child_adolescent/news_events/news/imnci-strategic-review/en/ (accessed on 1 June 2017).
- Feikin, D.R.; Njenga, M.K.; Bigogo, G.; Aura, B.; Aol, G.; Audi, A.; Jagero, G.; Muluare, P.O.; Gikunju, S.; Nderitu, L.; et al. Etiology and incidence of viral and bacterial acute respiratory illness among older children and adults in rural Western Kenya, 2007–2010. PLoS ONE 2012, 7, e43656. [Google Scholar] [CrossRef] [PubMed]
- Feikin, D.R.; Njenga, M.K.; Bigogo, G.; Aura, B.; Aol, G.; Audi, A.; Jagero, G.; Muluare, P.O.; Gikunju, S.; Nderitu, L.; et al. Viral and bacterial causes of severe acute respiratory illness among children aged less than 5 years in a high malaria prevalence area of Western Kenya, 2007–2010. Pediatr. Infect. Dis. J. 2013, 32, e14–e19. [Google Scholar] [CrossRef] [PubMed]
- Melamed, R.; Storch, G.A.; Holtz, L.R.; Klein, E.J.; Herrin, B.; Tarr, P.I.; Denno, D.M. Case-control assessment of the roles of noroviruses, human bocaviruses 2, 3 and 4, and novel polyomaviruses and astroviruses in acute childhood diarrhea. J. Pediatr. Infect. Dis. Soc. 2017. [Google Scholar] [CrossRef] [PubMed]
- WHO. Prequalification of in Vitro Diagnostics. Available online: http://www.who.int/diagnostics_laboratory/evaluations/en/ (accessed on 7 July 2017).
- FDA. FDA News Release. Available online: https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm543160.htm (accessed on 10 July 2017).
- Page, A.L.; de Rekeneire, N.; Sayadi, S.; Aberrane, S.; Janssens, A.C.; Dehoux, M.; Baron, E. Diagnostic and prognostic value of procalcitonin and c-reactive protein in malnourished children. Pediatrics 2014, 133, e363–e370. [Google Scholar] [CrossRef] [PubMed]
- Erdman, L.K.; D’Acremont, V.; Hayford, K.; Rajwans, N.; Kilowoko, M.; Kyungu, E.; Hongoa, P.; Alamo, L.; Streiner, D.L.; Genton, B.; et al. Biomarkers of host response predict primary end-point radiological pneumonia in Tanzanian children with clinical pneumonia: A prospective cohort study. PLoS ONE 2015, 10, e0137592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mancini, N.; Sambri, V.; Corti, C.; Ghidoli, N.; Tolomelli, G.; Paolucci, M.; Clerici, D.; Carletti, S.; Greco, R.; Tassara, M.; et al. Cost-effectiveness of blood culture and a multiplex real-time pcr in hematological patients with suspected sepsis: An observational propensity score-matched study. Expert Rev. Mol. Diagn. 2014, 14, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Valim, C.; Ahmad, R.; Lanaspa, M.; Tan, Y.; Acacio, S.; Gillette, M.A.; Almendinger, K.D.; Milner, D.A., Jr.; Madrid, L.; Pelle, K.; et al. Responses to bacteria, virus, and malaria distinguish the etiology of pediatric clinical pneumonia. Am. J. Respir. Crit. Care Med. 2016, 193, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.E.; Khatri, P. Generalizable biomarkers in critical care: Toward precision medicine. Crit. Care Med. 2017, 45, 934–939. [Google Scholar] [CrossRef] [PubMed]
- FIND. Samples & Reference Materials—Tuberculosis. Available online: https://www.finddx.org/specimen-banks/ (accessed on 7 July 2017).
- WHO. Integrated Management of Adolescent and Adult Illness (IMAI). Available online: http://www.who.int/3by5/publications/documents/imai/en/ or http://www.who.int/3by5/publications/documents/imai/en/ (accessed on 1 June 2017).
- Rambaud-Althaus, C.; Shao, A.F.; Kahama-Maro, J.; Genton, B.; d’Acremont, V. Managing the sick child in the era of declining malaria transmission: Development of almanach, an electronic algorithm for appropriate use of antimicrobials. PLoS ONE 2015, 10, e0127674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, A.F.; Rambaud-Althaus, C.; Samaka, J.; Faustine, A.F.; Perri-Moore, S.; Swai, N.; Kahama-Maro, J.; Mitchell, M.; Genton, B.; D’Acremont, V. New algorithm for managing childhood illness using mobile technology (almanach): A controlled non-inferiority study on clinical outcome and antibiotic use in Tanzania. PLoS ONE 2015, 10, e0132316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, A.F.; Rambaud-Althaus, C.; Swai, N.; Kahama-Maro, J.; Genton, B.; D’Acremont, V.; Pfeiffer, C. Can smartphones and tablets improve the management of childhood illness in Tanzania? A qualitative study from a primary health care worker’s perspective. BMC Health Serv. Res. 2015, 15, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keitel, K.; Kagoro, F.; Samaka, J.; Masimba, J.; Mlaganile, T.; Said, Z.; Temba, H.; Sangu, W.; Gervaix, A.; Genton, B.; et al. A Novel Electronic Algorithm Using Host Biomarker Point-of-Care-Tests for Management of Fever in under-Fives in Resource-Poor Settings (E-Poct): A Controlled, Non-Inferiority Study; Poster 481; ASTMH: Atlanta, GA, USA, 2016. [Google Scholar]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escadafal, C.; Nsanzabana, C.; Archer, J.; Chihota, V.; Rodriguez, W.; Dittrich, S. New Biomarkers and Diagnostic Tools for the Management of Fever in Low- and Middle-Income Countries: An Overview of the Challenges. Diagnostics 2017, 7, 44. https://doi.org/10.3390/diagnostics7030044
Escadafal C, Nsanzabana C, Archer J, Chihota V, Rodriguez W, Dittrich S. New Biomarkers and Diagnostic Tools for the Management of Fever in Low- and Middle-Income Countries: An Overview of the Challenges. Diagnostics. 2017; 7(3):44. https://doi.org/10.3390/diagnostics7030044
Chicago/Turabian StyleEscadafal, Camille, Christian Nsanzabana, Julie Archer, Violet Chihota, William Rodriguez, and Sabine Dittrich. 2017. "New Biomarkers and Diagnostic Tools for the Management of Fever in Low- and Middle-Income Countries: An Overview of the Challenges" Diagnostics 7, no. 3: 44. https://doi.org/10.3390/diagnostics7030044