Immortalized Muscle Cell Model to Test the Exon Skipping Efficacy for Duchenne Muscular Dystrophy
Abstract
:1. Introduction
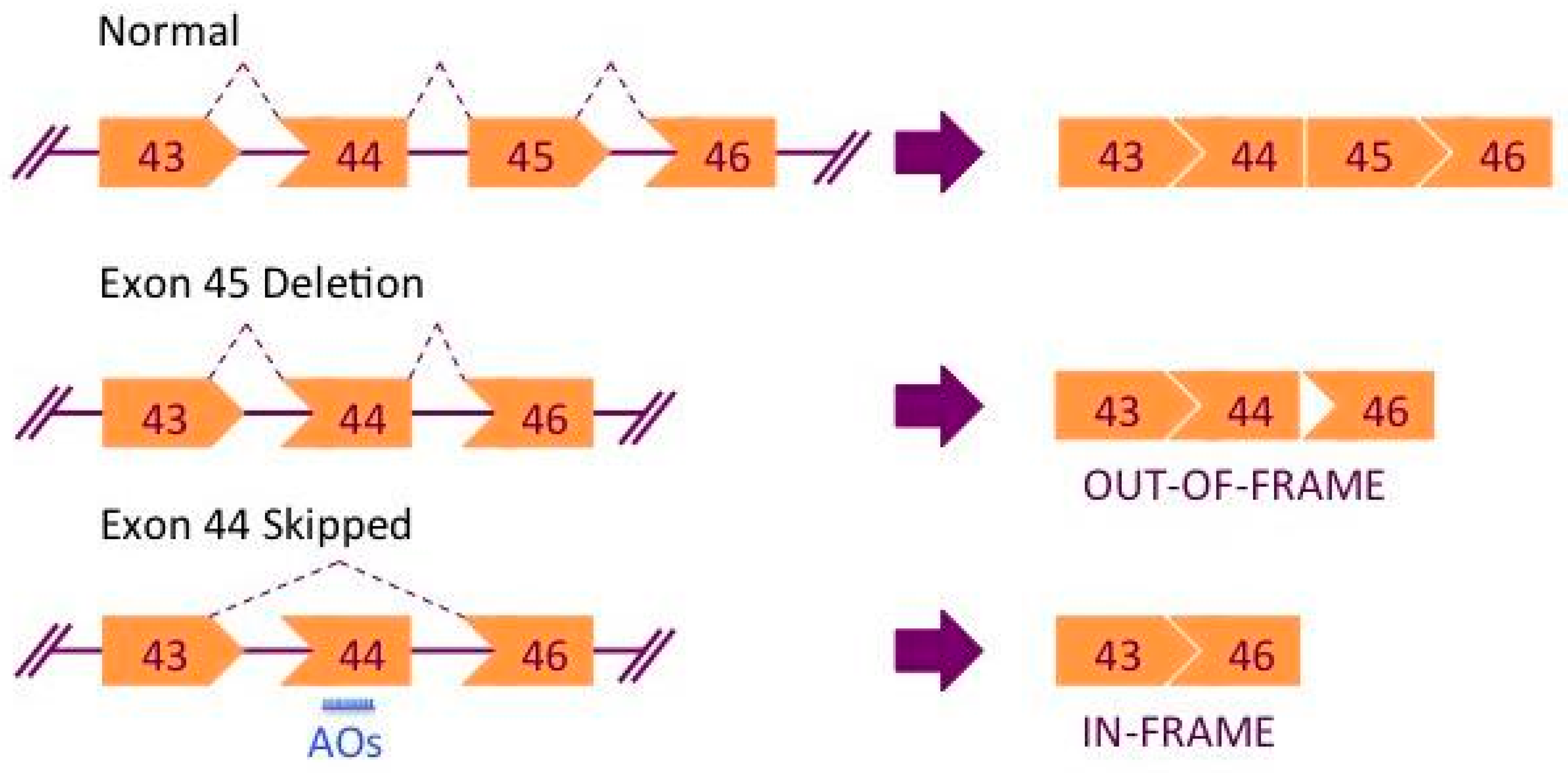
2. Exon Skipping Therapy for DMD
3. Challenges with Primary Cell Models for In Vitro Studies
4. Immortalized Cell Lines as a New Tool for In Vitro Studies
5. Considerations Regarding Immortalized Cell Lines
Acknowledgments
Conflicts of Interest
References
- Emery, A.E.H. Population frequencies of inherited neuromuscular diseases—A world survey. Neuromuscul. Disord. 1991, 1, 19–29. [Google Scholar] [CrossRef]
- Hoffman, E.P.; Brown, R.H.; Kunkel, L.M. Dystrophin: The protein product of the duchenne muscular dystrophy locus. Cell 1987, 51, 919–928. [Google Scholar] [CrossRef]
- Koenig, M.; Hoffman, E.P.; Bertelson, C.J.; Monaco, A.P.; Feener, C.; Kunkel, L.M. Complete cloning of the duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 1987, 50, 509–517. [Google Scholar] [CrossRef]
- Yokota, T.; Pistilli, E.; Duddy, W.; Nagaraju, K. Potential of oligonucleotide-mediated exon-skipping therapy for Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 2007, 7, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.P.; Kunkel, L.M.; Angelini, C.; Clarke, A.; Johnson, M.; Harris, J.B. Improved diagnosis of Becker muscular dystrophy by dystrophin testing. Neurology 1989, 39, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Arahata, K.; Hoffman, E.P.; Kunkel, L.M.; Ishiura, S.; Tsukahara, T.; Ishihara, T.; Sunohara, N.; Nonaka, I.; Ozawa, E.; Sugita, H. Dystrophin diagnosis: Comparison of dystrophin abnormalities by immunofluorescence and immunoblot analyses. Proc. Natl. Acad. Sci. USA 1989, 86, 7154–7158. [Google Scholar] [CrossRef] [PubMed]
- Neri, M.; Torelli, S.; Brown, S.; Ugo, I.; Sabatelli, P.; Merlini, L.; Spitali, P.; Rimessi, P.; Gualandi, F.; Sewry, C.; et al. Dystrophin levels as low as 30% are sufficient to avoid muscular dystrophy in the human. Neuromuscul. Disord. 2007, 17, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.P. Three muscular dystrophies: Loss of cytoskeleton-extracellular matrix linkage. Cell 1995, 80, 675–679. [Google Scholar] [CrossRef]
- Whitmore, C.; Morgan, J. What do mouse models of muscular dystrophy tell us about the DAPC and its components? Int. J. Exp. Pathol. 2014, 95, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Sunada, Y.; Campbell, K.P. Dystrophin-glycoprotein complex: Molecular organization and critical roles in skeletal muscle. Curr. Opin. Neurol. 1995, 8, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Nichols, B.; Takeda, S.; Yokota, T. Nonmechanical Roles of Dystrophin and Associated Proteins in Exercise, Neuromuscular Junctions, and Brains. Brain Sci. 2015, 5, 275–298. [Google Scholar] [CrossRef] [PubMed]
- Lapidos, K.A.; Kakkar, R.; McNally, E.M. The Dystrophin Glycoprotein Complex: Signaling Strength and Integrity for the Sarcolemma. Circ. Res. 2004, 94, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Takeda, S. Mammalian models of duchenne muscular dystrophy: Pathological characteristics and therapeutic applications. J. Biomed. Biotechnol. 2011, 2011, 184393. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.W.; Whitehead, N.P.; Suchyna, T.M.; Gottlieb, P.A.; Sachs, F.; Allen, D.G. Effects of stretch-activated channel blockers on [Ca2+]i and muscle damage in the mdx mouse. J. Physiol. 2005, 562, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.D. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum. Mol. Genet. 2002, 11, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Moser, H. Duchenne muscular dystrophy: Pathogenetic aspects and genetic prevention. Hum. Genet. 1984, 66, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Duchenne, G.-B.-A. The pathology of paralysis with muscular degeneration (Paralysie Myosclerotique), or paralysis with apparent hypertrophy. Br. Med. J. 1867, 2, 541–542. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.F.; Kunkel, L.M. Dystrophin Abnormalities in Duchenne/Becker Muscular Review Dystrophy genes. Cell 1989, 2, 1019–1029. [Google Scholar]
- Eagle, M.; Baudouin, S.V.; Chandler, C.; Giddings, D.R.; Bullock, R.; Bushby, K. Survival in Duchenne muscular dystrophy: Improvements in life expectancy since 1967 and the impact of home nocturnal ventilation. Neuromuscul. Disord. 2002, 12, 926–929. [Google Scholar] [CrossRef]
- Passamano, L.; Taglia, A.; Palladino, A.; Viggiano, E.; D’Ambrosio, P.; Scutifero, M.; Cecio, M.R.; Torre, V.; De Luca, F.; Picillo, E.; et al. Improvement of survival in Duchenne Muscular Dystrophy: Retrospective analysis of 835 patients. Acta Myol. 2012, 31, 121–125. [Google Scholar] [PubMed]
- Pandey, S.N.; Kesari, A.; Yokota, T.; Pandey, G.S. Muscular dystrophy: Disease mechanisms and therapies. BioMed Res. Int. 2015, 2015, 456348. [Google Scholar] [CrossRef] [PubMed]
- Touznik, A.; Lee, J.J.A.; Yokota, T. New developments in exon skipping and splice modulation therapies for neuromuscular diseases. Expert Opin. Biol. Ther. 2014, 14, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yokota, T. Antisense Therapy in Neurology. J. Pers. Med. 2013, 3, 144–176. [Google Scholar] [CrossRef] [PubMed]
- Kuzmiak, H.A.; Maquat, L.E. Applying nonsense-mediated mRNA decay research to the clinic: Progress and challenges. Trends Mol. Med. 2006, 12, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.A.; Yokota, T. Translational Research in Nucleic Acid Therapies for Muscular Dystrophies. In Translational Research in Muscular Dystrophy; Springer: Tokyo, Japan, 2016; pp. 87–102. [Google Scholar]
- Aoki, Y.; Nakamura, A.; Yokota, T.; Saito, T.; Okazawa, H.; Nagata, T.; Takeda, S. In-frame Dystrophin Following Exon 51-Skipping Improves Muscle Pathology and Function in the Exon 52–Deficient mdx Mouse. Mol. Ther. 2010, 18, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Duddy, W.; Echigoya, Y.; Kolski, H. Exon skipping for nonsense mutations in Duchenne muscular dystrophy: Too many mutations, too few patients? Expert Opin. Biol. Ther. 2012, 12, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, R.; Echigoya, Y.; Caluseriu, O.; Aoki, Y.; Takeda, S.; Yokota, T. Systemic delivery of morpholinos to skip multiple exons in a dog model of duchenne muscular dystrophy. In Methods in Molecular Biology; Moulton, H., Moulton, J., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1565, pp. 201–213. ISBN 978-1-4939-6817-6. [Google Scholar]
- Wein, N.; Vulin, A.; Findlay, A.R.; Gumienny, F.; Huang, N.; Wilton, S.D.; Flanigan, K.M. Efficient Skipping of Single Exon Duplications in DMD Patient-Derived Cell Lines Using an Antisense Oligonucleotide Approach. J. Neuromuscul. Dis. 2017, 4, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Echigoya, Y.; Fukada, S.-I.; Yokota, T. Current Translational Research and Murine Models For Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2016, 3, 29–48. [Google Scholar] [CrossRef] [PubMed]
- McGreevy, J.W.; Hakim, C.H.; McIntosh, M.A.; Duan, D. Animal models of Duchenne muscular dystrophy: From basic mechanisms to gene therapy. Dis. Model. Mech. 2015, 8, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Bao, B.; Echigoya, Y.; Yokota, T. Dystrophin-deficient large animal models: Translational research and exon skipping. Am. J. Transl. Res. 2015, 7, 1314–1331. [Google Scholar] [PubMed]
- Echigoya, Y.; Lim, K.R.Q.; Trieu, N.; Bao, B.; Miskew, B.; Vila, M.C.; Novak, J.S.; Hara, Y.; Lee, J.; Touznik, A.; et al. Quantitative antisense screening and optimization for exon 51 skipping in Duchenne muscular dystrophy. Mol. Ther. 2017, 25. [Google Scholar] [CrossRef] [PubMed]
- Young, C.S.; Mokhonova, E.; Quinonez, M.; Pyle, A.D.; Spencer, M.J. Creation of a Novel Humanized Dystrophic Mouse Model of Duchenne Muscular Dystrophy and Application of a CRISPR/Cas9 Gene Editing Therapy. J. Neuromuscul. Dis. 2017, 4, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; Janson, A.A.M.; Kaman, W.E.; Bremmer-Bout, M.; den Dunnen, J.T.; Baas, F.; van Ommen, G.J.B.; van Deutekom, J.C.T. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum. Mol. Genet. 2003, 12, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Arechavala-Gomeza, V.; Graham, I.R.; Popplewell, L.J.; Adams, A.M.; Aartsma-Rus, A.; Kinali, M.; Morgan, J.E.; van Deutekom, J.C.; Wilton, S.D.; Dickson, G.; et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum. Gene Ther. 2007, 18, 798–810. [Google Scholar] [CrossRef] [PubMed]
- Renault, V.; Thorne, L.-E.; Eriksson, P.-O.; Butler-Browne, G.; Mouly, V. Regenerative potential of human skeletal muscle during aging. Aging Cell 2002, 1, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Wright, W.E.; Shay, J.W. Historical claims and current interpretations of replicative aging. Nat. Biotechnol. 2002, 20, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.; Blau, H.M. Accelerated age-related decline in replicative life-span of Duchenne muscular dystrophy myoblasts: Implications for cell and gene therapy. Somat. Cell Mol. Genet. 1990, 16, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Mamchaoui, K.; Trollet, C.; Bigot, A.; Negroni, E.; Chaouch, S.; Wolff, A.; Kandalla, P.K.; Marie, S.; Di Santo, J.; St Guily, J.; et al. Immortalized pathological human myoblasts: Towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle 2011, 1, 34. [Google Scholar] [CrossRef] [PubMed]
- Périé, S.; Mamchaoui, K.; Mouly, V.; Blot, S.; Bouazza, B.; Thornell, L.E.; St Guily, J.L.; Butler-Browne, G. Premature proliferative arrest of cricopharyngeal myoblasts in oculo-pharyngeal muscular dystrophy: Therapeutic perspectives of autologous myoblast transplantation. Neuromuscul. Disord. 2006, 16, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; Bremmer-Bout, M.; Janson, A.A.M.; Den Dunnen, J.T.; Van Ommen, G.J.B.; Van Deutekom, J.C.T. Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscul. Disord. 2002, 12, S71–S77. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; De winter, C.L.; Janson, A.A.M.; Kaman, W.E.; Van ommen, G.B.; Den dunnen, J.T.; Van deutekom, J.C.T. Functional Analysis of 114 Exon-Internal AONs for Targeted DMD Exon Skipping: Indication for Steric Hindrance of SR Protein Binding Sites. Oligonucleotides 2005, 15, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; Janson, A.A.; van Ommen, G.-J.B.; van Deutekom, J.C. Antisense-induced exon skipping for duplications in Duchenne muscular dystrophy. BMC Med. Genet. 2007, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Popplewell, L.J.; Adkin, C.; Arechavala-Gomeza, V.; Aartsma-Rus, A.; de Winter, C.L.; Wilton, S.D.; Morgan, J.E.; Muntoni, F.; Graham, I.R.; Dickson, G. Comparative analysis of antisense oligonucleotide sequences targeting exon 53 of the human DMD gene: Implications for future clinical trials. Neuromuscul. Disord. 2010, 20, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Shimo, T.; Tachibana, K.; Saito, K.; Yoshida, T.; Tomita, E.; Waki, R.; Yamamoto, T.; Doi, T.; Inoue, T.; Kawakami, J.; et al. Design and evaluation of locked nucleic acid-based splice-switching oligonucleotides in vitro. Nucleic Acids Res. 2014, 42, 8174–8187. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Nakamura, A.; Aoki, Y.; Yokota, T.; Okada, T.; Osawa, M.; Takeda, S. Antisense PMO found in Dystrophic Dog model was effective in cells from exon 7-deleted DMD patient. PLoS ONE 2010, 5, e12239. [Google Scholar] [CrossRef] [PubMed]
- Echigoya, Y.; Mouly, V.; Garcia, L.; Yokota, T.; Duddy, W. In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in duchenne muscular dystrophy. PLoS ONE 2015, 10, e0120058. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.H.; Mouly, V.; Cooper, R.N.; Mamchaoui, K.; Bigot, A.; Shay, J.W.; Di Santo, J.P.; Butler-Browne, G.S.; Wright, W.E. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: Consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell 2007, 6, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Thorley, M.; Duguez, S.; Mazza, E.M.C.; Valsoni, S.; Bigot, A.; Mamchaoui, K.; Harmon, B.; Voit, T.; Mouly, V.; Duddy, W. Skeletal muscle characteristics are preserved in hTERT/cdk4 human myogenic cell lines. Skelet. Muscle 2016, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- The Lancet. Patient need versus evidence: A balancing act. Lancet 2016, 388, 1350. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Krieg, A.M. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Ther. 2017, 27, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.R.Q.; Maruyama, R.; Yokota, T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Dev. Ther. 2017, 11, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Kesselheim, A.S.; Avorn, J. Approving a Problematic Muscular Dystrophy Drug. JAMA 2016, 316, 2357–2358. [Google Scholar] [CrossRef] [PubMed]
- Voit, T.; Topaloglu, H.; Straub, V.; Muntoni, F.; Deconinck, N.; Campion, G.; De Kimpe, S.J.; Eagle, M.; Guglieri, M.; Hood, S.; et al. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): An exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014, 13, 987–996. [Google Scholar] [CrossRef]
- Guncay, A.; Yokota, T. Antisense oligonucleotide drugs for Duchenne muscular dystrophy: How far have we come and what does the future hold? Future Med. Chem. 2015, 7, 1631–1635. [Google Scholar] [CrossRef] [PubMed]
- Chaouch, S.; Mouly, V.; Goyenvalle, A.; Vulin, A.; Mamchaoui, K.; Negroni, E.; Di Santo, J.; Butler-Browne, G.; Torrente, Y.; Garcia, L.; et al. Immortalized Skin Fibroblasts Expressing Conditional MyoD as a Renewable and Reliable Source of Converted Human Muscle Cells to Assess Therapeutic Strategies for Muscular Dystrophies: Validation of an Exon-Skipping Approach to Restore Dystrophin in Duchen. Hum. Gene Ther. 2009, 20, 784–790. [Google Scholar] [CrossRef] [PubMed]
- González-Suárez, E.; Samper, E.; Ramírez, A.; Flores, J.M.; Martín-Caballero, J.; Jorcano, J.L.; Blasco, M.A. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001, 20, 2619–2630. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.A.; Hahn, W.C.; O’Connor, B.F.; Banner, E.N.; Lundberg, A.S.; Modha, P.; Mizuno, H.; Brooks, M.W.; Fleming, M.; Zimonjic, D.B.; et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. USA 2002, 99, 12606–12611. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.L.; Coller, H.A.; Roberts, J.M. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003, 5, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-I.; Venteicher, A.S.; Hong, J.Y.; Choi, J.; Jun, S.; Shkreli, M.; Chang, W.; Meng, Z.; Cheung, P.; Ji, H.; et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009, 460, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Tsivitse, S. Notch and Wnt signaling, physiological stimuli and postnatal myogenesis. Int. J. Biol. Sci. 2010, 6, 268–281. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, Q.; Yokota, T. Immortalized Muscle Cell Model to Test the Exon Skipping Efficacy for Duchenne Muscular Dystrophy. J. Pers. Med. 2017, 7, 13. https://doi.org/10.3390/jpm7040013
Nguyen Q, Yokota T. Immortalized Muscle Cell Model to Test the Exon Skipping Efficacy for Duchenne Muscular Dystrophy. Journal of Personalized Medicine. 2017; 7(4):13. https://doi.org/10.3390/jpm7040013
Chicago/Turabian StyleNguyen, Quynh, and Toshifumi Yokota. 2017. "Immortalized Muscle Cell Model to Test the Exon Skipping Efficacy for Duchenne Muscular Dystrophy" Journal of Personalized Medicine 7, no. 4: 13. https://doi.org/10.3390/jpm7040013






