Tailoring Nutritional Advice for Mexicans Based on Prevalence Profiles of Diet-Related Adaptive Gene Polymorphisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genotyping
2.3. Copy Number Variation
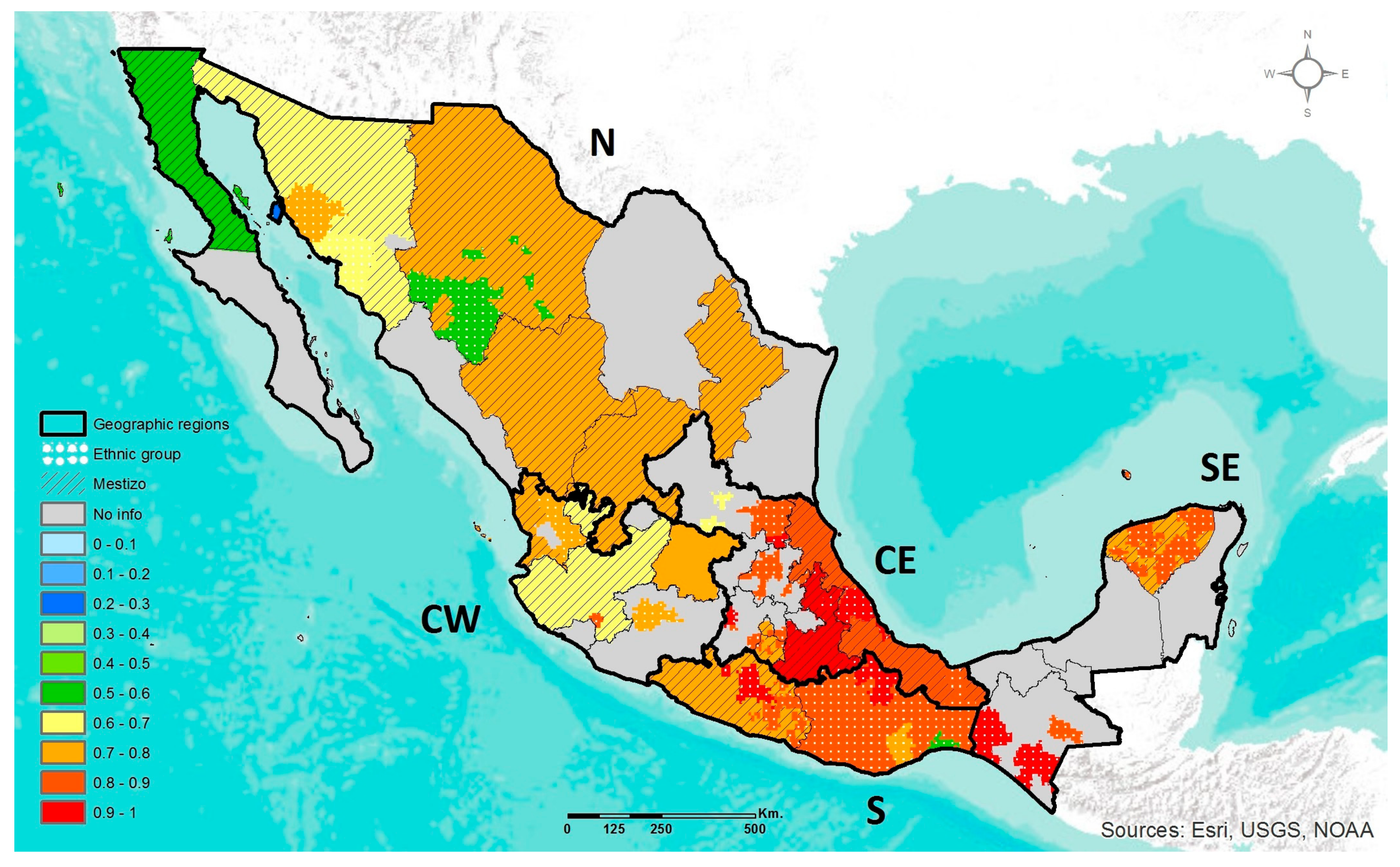
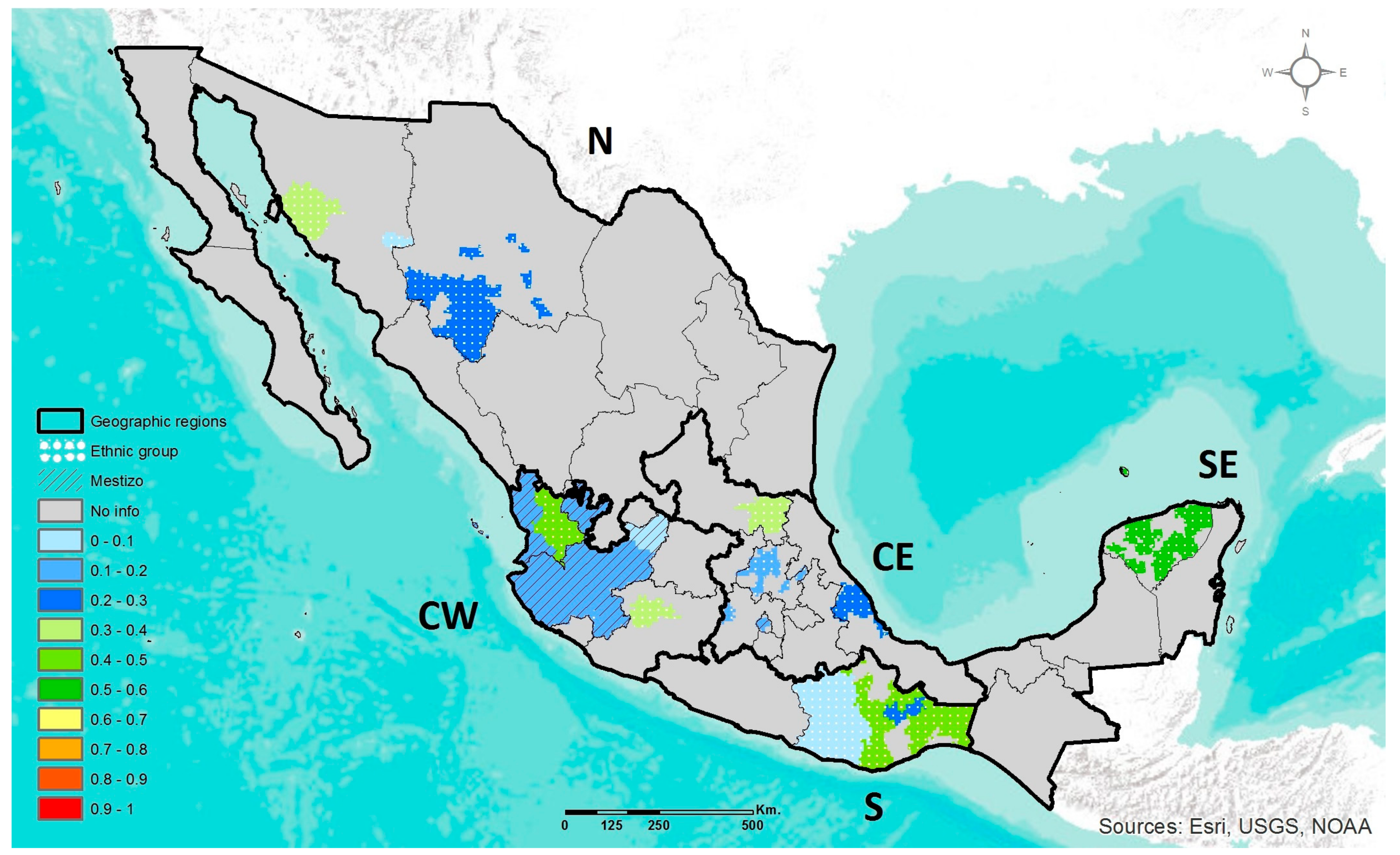
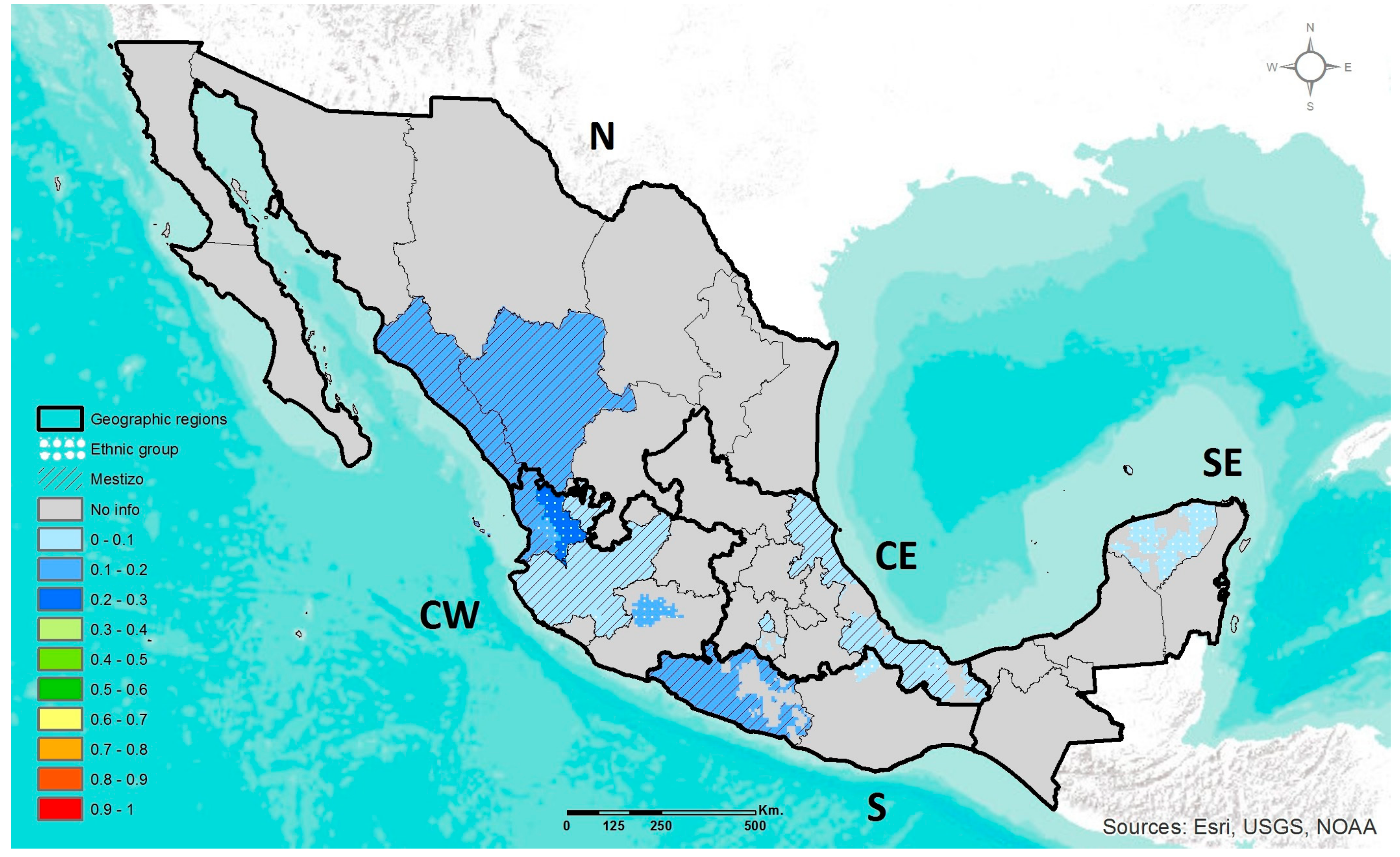
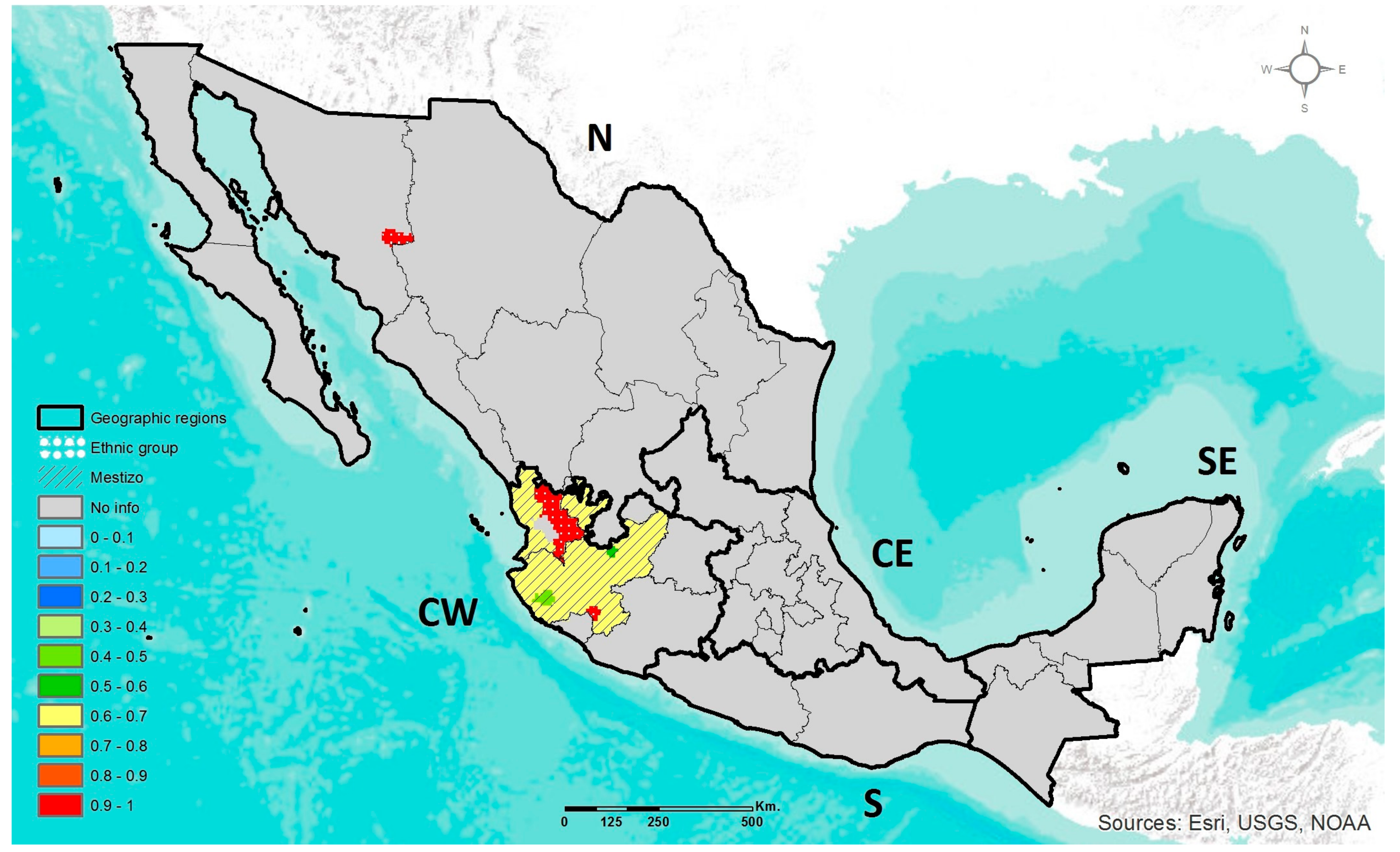
2.4. Geographic Heat Maps
2.5. Statistical Analysis
2.6. Ethical Guidelines
3. Results
3.1. Prevalence of DRAG Polymorphisms in Mexico
3.1.1. MTHFR C677T
3.1.2. ABCA1 R230C
3.1.3. APOE ε2, ε3, ε4
3.1.4. LCT C-13910T
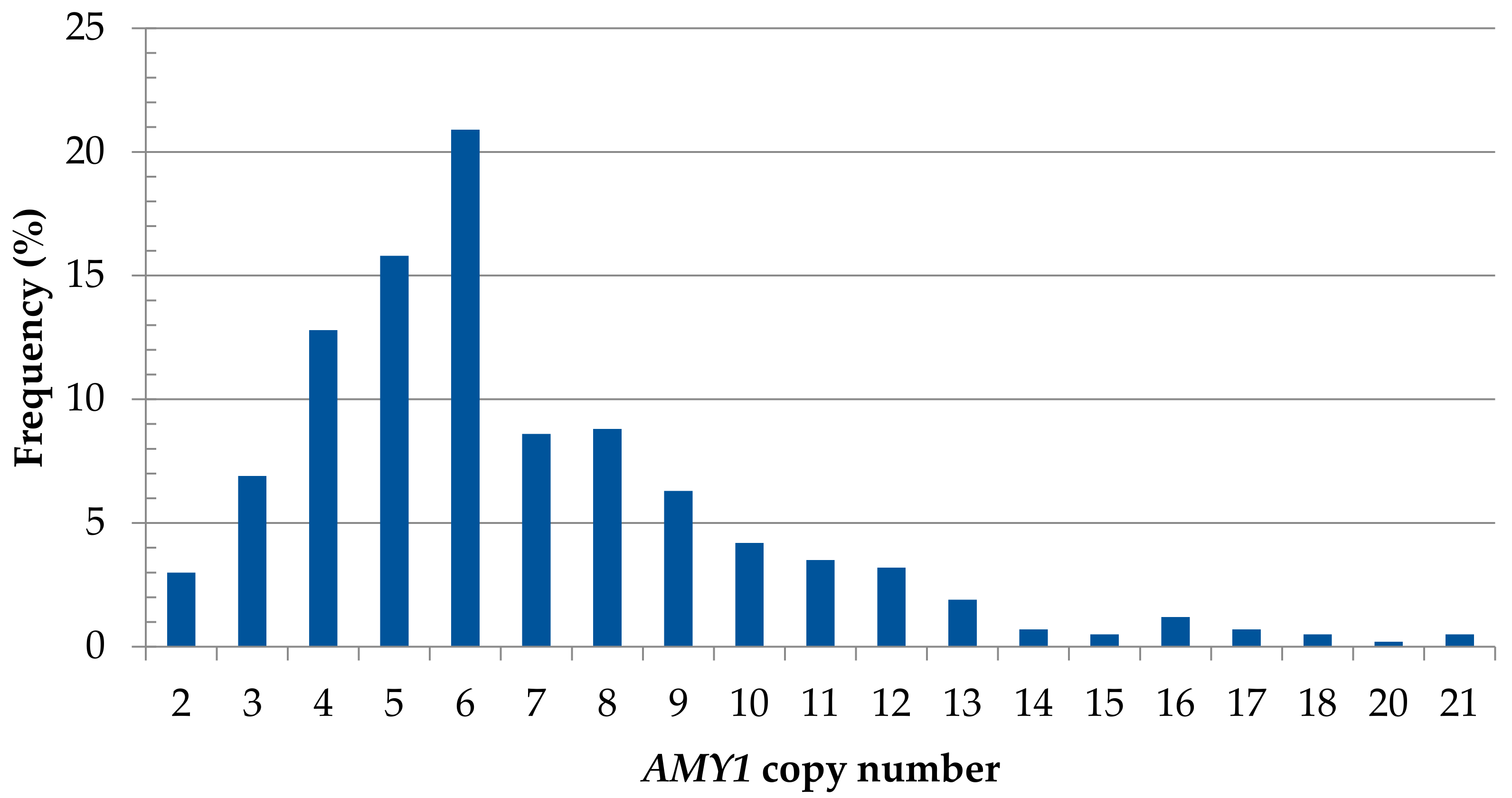
3.1.5. AMY1 Copy Number Variation
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Panduro, A.; Vázquez-Castellanos, J.L. Genes y medio ambiente. Investig. Salud 2001, 3, 41–48. [Google Scholar]
- Popkin, B.M.; Adair, L.S.; Ng, S.W. Global nutrition transition and the pandemic of obesity in developing countries. Nutr. Rev. 2012, 70, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Kaput, J. Diet-disease gene interactions. Nutrition 2004, 20, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Hansen, M.E.; Lo, Y.; Tishkoff, S.A. Going global by adapting local: A review of recent human adaptation. Science 2016, 354, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, G.P. Why Some Like It Hot: Food, Genes, and Cultural Diversity; Island Press/Shearwater Books: Washington, DC, USA, 2004; p. 233. [Google Scholar]
- Chiaroni, J.; Underhill, P.A.; Cavalli-Sforza, L.L. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 20174–20179. [Google Scholar] [CrossRef] [PubMed]
- Laland, K.N.; Odling-Smee, J.; Myles, S. How culture shaped the human genome: Bringing genetics and the human sciences together. Nat. Rev. Genet. 2010, 11, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Hancock, A.M.; Witonsky, D.B.; Ehler, E.; Alkorta-Aranburu, G.; Beall, C.; Gebremedhin, A.; Sukernik, R.; Utermann, G.; Pritchard, J.; Coop, G.; et al. Colloquium paper: Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. S2), 8924–8930. [Google Scholar] [CrossRef] [PubMed]
- Babbitt, C.C.; Warner, L.R.; Fedrigo, O.; Wall, C.E.; Wray, G.A. Genomic signatures of diet-related shifts during human origins. Proc. Biol. Sci. 2011, 278, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Bersaglieri, T.; Sabeti, P.C.; Patterson, N.; Vanderploeg, T.; Schaffner, S.F.; Drake, J.A.; Rhodes, M.; Reich, D.E.; Hirschhorn, J.N. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004, 74, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Ingram, C.J.; Elamin, M.F.; Mulcare, C.A.; Weale, M.E.; Tarekegn, A.; Raga, T.O.; Bekele, E.; Elamin, F.M.; Thomas, M.G.; Bradman, N.; et al. A novel polymorphism associated with lactose tolerance in Africa: Multiple causes for lactase persistence? Hum. Genet. 2007, 120, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Perry, G.H.; Dominy, N.J.; Claw, K.G.; Lee, A.S.; Fiegler, H.; Redon, R.; Werner, J.; Villanea, F.A.; Mountain, J.L.; Misra, R.; et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007, 39, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Ko, A.; Cantor, R.M.; Weissglas-Volkov, D.; Nikkola, E.; Reddy, P.M.; Sinsheimer, J.S.; Pasaniuc, B.; Brown, R.; Alvarez, M.; Rodriguez, A.; et al. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat. Commun. 2014, 5, 3983. [Google Scholar] [CrossRef] [PubMed]
- De Montellano, B.O. Aztec Medicine, Health, and Nutrition; Rutgers University Press: New Brunswick, NJ, USA, 1990; p. 308. [Google Scholar]
- Zizumbo-Villarreal, D.; Flores-Silva, A.; Colunga-García, P. The archaic diet in Mesoamerica: Incentive for milpa development and species domestication. Econom. Bot. 2012, 66, 328–343. [Google Scholar] [CrossRef]
- Kraft, K.H.; Brown, C.H.; Nabhan, G.P.; Luedeling, E.; Luna Ruiz Jde, J.; Coppens d’Eeckenbrugge, G.; Hijmans, R.J.; Gepts, P. Multiple lines of evidence for the origin of domesticated chili pepper, Capsicum annuum, in Mexico. Proc. Natl. Acad. Sci. USA 2014, 111, 6165–6170. [Google Scholar] [CrossRef] [PubMed]
- Almaguer González, J.A.; García Ramirez, H.J. La Dieta de la Milpa. Modelo de Alimentación Mesoamericana Saludable y Culturalmente Saludable; Secretaría de Salud. Available online: https://www.gob.mx/salud/acciones-y-programas/la-dieta-de-la-milpa (Assessed on 30 October 2017).
- Mutchinick, O.M.; López, M.A.; Luna, L.; Waxman, J.; Babinsky, V.E. High prevalence of the thermolabile methylenetetrahydrofolate reductase variant in Mexico: A country with a very high prevalence of neural tube defects. Mol. Genet. Metab. 1999, 68, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Hünemeier, T.; Amorim, C.E.; Azevedo, S.; Contini, V.; Acuña-Alonzo, V.; Rothhammer, F.; Dugoujon, J.M.; Mazières, S.; Barrantes, R.; Villarreal-Molina, M.T.; et al. Evolutionary responses to a constructed niche: Ancient Mesoamericans as a model of gene-culture coevolution. PLoS ONE 2012, 7, e38862. [Google Scholar] [CrossRef]
- Aceves, D.; Ruiz, B.; Nuño, P.; Roman, S.; Zepeda, E.; Panduro, A. Heterogeneity of apolipoprotein E polymorphism in different Mexican populations. Hum. Biol. 2006, 78, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Estrada, A.; Gignoux, C.R.; Fernández-Lopez, J.C.; Zakharia, F.; Sikora, M.; Contreras, A.V.; Acuña-Alonzo, V.; Sandoval, K.; Eng, C.; Romero-Hidalgo, S.; et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 2014, 344, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Villalobos, H.; Muñoz-Valle, J.F.; González-Martín, A.; Gorostiza, A.; Magaña, M.T.; Páez-Riberos, L.A. Genetic admixture, relatedness, and structure patterns among Mexican populations revealed by the Y-chromosome. Am. J. Phys. Anthropol. 2008, 135, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Aldaco, K.; Rebello Pinho, J.R.; Roman, S.; Gleyzer, K.; Fierro, N.A.; Oyakawa, L.; Ramos-Lopez, O.; Ferraz Santana, R.A.; Sitnik, R.; Panduro, A. Association with spontaneous hepatitis C viral clearance and genetic differentiation of IL28B/IFNL4 haplotypes in populations from Mexico. PLoS ONE 2016, 11, e0146258. [Google Scholar] [CrossRef] [PubMed]
- Ojeda-Granados, C.; Panduro, A.; Rebello Pinho, J.R.; Ramos-Lopez, O.; Gleyzer, K.; Malta, F.M.; Gonzalez-Aldaco, K.; Roman, S. Association of lactase persistence genotypes with high intake of dairy saturated fat and high prevalence of lactase non-persistence among the Mexican population. J. Nutrigenet. Nutrigenom. 2016, 9, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [PubMed]
- Mejia-Benítez, M.A.; Bonnefond, A.; Yengo, L.; Huyvaert, M.; Dechaume, A.; Peralta-Romero, J.; Klünder-Klünder, M.; García Mena, J.; El-Sayed Moustafa, J.S.; Falchi, M.; et al. Beneficial effect of a high number of copies of salivary amylase AMY1 gene on obesity risk in Mexican children. Diabetologia 2015, 58, 290–294. [Google Scholar] [CrossRef]
- Garcia, L.; Gold, E.B.; Wang, L.; Yang, X.; Mao, M.; Schwartz, A.V. The relation of acculturation to overweight, obesity, pre-diabetes and diabetes among U.S. Mexican-American women and men. Ethn. Dis. 2012, 22, 58–64. [Google Scholar] [PubMed]
- Kuhnlein, H.V.; Receveur, O.; Soueida, R.; Egeland, G.M. Arctic indigenous peoples experience the nutrition transition with changing dietary patterns and obesity. J. Nutr. 2004, 134, 1447–1453. [Google Scholar] [PubMed]
- Roman, S.; Ojeda-Granados, C.; Panduro, A. Genética y evolución de la alimentación de la población en México. Rev. Endocrinol. Nutr. 2013, 21, 42–51. [Google Scholar]
- Guéant-Rodriguez, R.M.; Guéant, J.L.; Debard, R.; Thirion, S.; Hong, L.X.; Bronowicki, J.P.; Namour, F.; Chabi, N.W.; Sanni, A.; Anello, G.; et al. Prevalence of methylenetetrahydrofolate reductase 677T and 1298C alleles and folate status: A comparative sudy in Mexican, West African, and European populations. Am. J. Clin. Nutr. 2006, 83, 701–707. [Google Scholar]
- Mayor-Olea, A.; Callejón, G.; Palomares, A.R.; Jiménez, A.J.; Gaitán, M.J.; Rodríguez, A.; Ruiz, M.; Reyes-Engel, A. Human genetic selection on the MTHFR 677C>T polymorphism. BMC Med. Genet. 2008, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Isordia-Salas, I.; Barinagarrementería-Aldatz, F.; Leaños-Miranda, A.; Borrayo-Sánchez, G.; Vela-Ojeda, J.; García-Chávez, J.; Ibarra-Gonzalez, I.; Majluf-Cruz, A. The C677T polymorphism of the methylenetetrahydrofolate reductase gene is associated with idiopathic ischemic stroke in the young Mexican-mestizo population. Cerebrovasc. Dis. 2010, 29, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Alaniz, F.; Lumbreras-Márquez, M.I.; Sandoval-Carrillo, A.A.; Aguilar-Durán, M.; Méndez-Hernández, E.M.; Barraza-Salas, M.; Castellanos-Juárez, F.X.; Salas-Pacheco, J.M. Association of COMT G675A and MTHFR C677T polymorphisms with hypertensive disorders of pregnancy in Mexican mestizo population. Preg. Hypertens. 2014, 4, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Gallegos-Arreola, M.P.; García-Ortiz, J.E.; Figuera, L.E.; Puebla-Pérez, A.M.; Morgan-Villela, G.; Zúñiga-González, G.M. Association of the 677C → T polymorphism in the MTHFR gene with colorectal cancer in Mexican patients. Cancer Genom. Proteom. 2009, 6, 183–188. [Google Scholar]
- Ramos-Silva, A.; Figuera, L.E.; Soto-Quintana, O.M.; Puebla-Pérez, A.M.; Ramirez-Patiño, R.; Gutiérrez-Hurtado, I.; Carrillo-Moreno, D.I.; Zúñiga-González, G.M.; Dávalos-Rodríguez, I.P.; Gallegos-Arreola, M.P. Association of the C677T polymorphism in the methylenetetrahydrofolate reductase gene with breast cancer in a Mexican population. Genet. Mol. Res. 2015, 14, 4015–4026. [Google Scholar] [CrossRef] [PubMed]
- Sazci, A.; Ergul, E.; Aygun, C.; Akpinar, G.; Senturk, O.; Hulagu, S. Methylenetetrahydrofolate reductase gene polymorphisms in patients with nonalcoholic steatohepatitis (NASH). Cell Biochem. Funct. 2008, 26, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Solis, C.; Veenema, K.; Ivanov, A.A.; Tran, S.; Li, R.; Wang, W.; Moriarty, D.J.; Maletz, C.V.; Caudill, M.A. Folate intake at RDA levels is inadequate for Mexican American men with the methylenetetrahydrofolate reductase 677TT genotype. J. Nutr. 2008, 138, 67–72. [Google Scholar] [PubMed]
- Ashfield-Watt, P.A.; Pullin, C.H.; Whiting, J.M.; Clark, Z.E.; Moat, S.J.; Newcombe, R.G.; Burr, M.L.; Lewis, M.J.; Powers, H.J.; McDowell, I.F. Methylenetetrahydrofolate reductase 677C → T genotype modulates homocysteine responses to a folate-rich diet or a low-dose folic acid supplement: A randomized controlled trial. Am. J. Clin. Nutr. 2002, 76, 180–186. [Google Scholar] [PubMed]
- Hung, J.; Yang, T.L.; Urrutia, T.F.; Li, R.; Perry, C.A.; Hata, H.; Cogger, E.A.; Moriarty, D.J.; Caudill, M.A. Additional food folate derived exclusively from natural sources improves folate status in young women with the MTHFR 677 CC or TT genotype. J. Nutr. Biochem. 2006, 17, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Villarreal-Molina, M.T.; Aguilar-Salinas, C.A.; Rodríguez-Cruz, M.; Riaño, D.; Villalobos-Comparan, M.; Coral-Vazquez, R.; Menjivar, M.; Yescas-Gomez, P.; Königsoerg-Fainstein, M.; Romero-Hidalgo, S.; et al. The ATP-binding cassette transporter A1 R230C variant affects HDL cholesterol levels and BMI in the Mexican population: Association with obesity and obesity-related comorbidities. Diabetes 2007, 56, 1881–1887. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, R.; Vargas-Alarcón, G.; Medina-Urrutia, A.; Cardoso-Saldaña, G.; Hernández-Pacheco, G.; Zamora-González, J.; Posadas-Romero, C. Influence of the apolipoprotein E polymorphism on plasma lipoproteins in a Mexican population. Hum. Biol. 2001, 73, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Acuña-Alonzo, V.; Flores-Dorantes, T.; Kruit, J.K.; Villarreal-Molina, T.; Arellano-Campos, O.; Hünemeier, T.; Moreno-Estrada, A.; Ortiz-López, M.G.; Villamil-Ramírez, H.; León-Mimila, P.; et al. A functional ABCA1 gene variant is associated with low HDL-cholesterol levels and shows evidence of positive selection in Native Americans. Hum. Mol. Genet. 2010, 19, 2877–2885. [Google Scholar] [CrossRef]
- Corbo, R.M.; Scacchi, R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele? Ann. Hum. Genet. 1999, 63, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Ramos-López, O.; Roman, S.; Ojeda-Granados, C.; Sepúlveda-Villegas, M.; Martínez-López, E.; Torres-Valadez, R.; Trujillo-Trujillo, E.; Panduro, A. Patrón de ingesta alimentaria y actividad física en pacientes hepatópatas en el occidente de méxico. Rev. Endocrinol. Nutr. 2013, 21, 7–15. [Google Scholar]
- Campos-Pérez, W.; González-Becerra, K.; Ramos-López, O.; Silva-Gómez, J.A.; Barrón-Cabrera, E.; Roman, S.; Panduro, A.; Martínez-López, E. Same dietary but different physical activity pattern in normal-weight and overweight Mexican subjects. J. Food Nutr. Res. 2016, 4, 729–735. [Google Scholar] [CrossRef]
- Sullivan, S. Implications of diet on nonalcoholic fatty liver disease. Curr. Opin. Gastroenterol. 2010, 26, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Ferolla, S.M.; Ferrari, T.C.; Lima, M.L.; Reis, T.O.; Tavares, W.C., Jr.; Couto, O.F.; Vidigal, P.V.; Fausto, M.A.; Couto, C.A. Dietary patterns in Brazilian patients with non-alcoholic fatty liver disease: A cross-sectional study. Clinics (Sao Paulo) 2013, 68, 11–17. [Google Scholar] [CrossRef]
- Villarreal-Molina, M.T.; Flores-Dorantes, M.T.; Arellano-Campos, O.; Villalobos-Comparan, M.; Rodríguez-Cruz, M.; Miliar-García, A.; Huertas-Vazquez, A.; Menjivar, M.; Romero-Hidalgo, S.; Wacher, N.H.; et al. Association of the ATP-binding cassette transporter A1 R230C variant with early-onset type 2 diabetes in a Mexican population. Diabetes 2008, 57, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Salinas, C.A.; Canizales-Quinteros, S.; Rojas-Martínez, R.; Mehta, R.; Rodriguez-Guillén, R.; Ordoñez-Sanchez, M.L.; Riba, L.; Tusié-Luna, M.T. The non-synonymous Arg230cCys variant (R230C) of the ATP-binding cassette transporter A1 is associated with low HDL cholesterol concentrations in Mexican adults: A population based nation wide study. Atherosclerosis 2011, 216, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Cahua-Pablo, G.; Cruz, M.; Moral-Hernández, O.D.; Leyva-Vázquez, M.A.; Antúnez-Ortiz, D.L.; Cahua-Pablo, J.A.; Alarcón-Romero Ldel, C.; Ortuño-Pineda, C.; Moreno-Godínez, M.E.; Hernández-Sotelo, D.; et al. Elevated levels of LDL-C are associated with ApoE4 but not with the rs688 polymorphism in the LDLR gene. Clin. Appl. Thromb. Hemost. 2016, 22, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, E.; Curiel-Lopez, F.; Hernandez-Nazara, A.; Moreno-Luna, L.E.; Ramos-Marquez, M.E.; Roman, S.; Panduro, A. Influence of APOE and FABP2 polymorphisms and environmental factors in the susceptibility to gallstone disease. Ann. Hepatol. 2015, 14, 515–523. [Google Scholar] [PubMed]
- Santos, A.; Salguero, M.L.; Gurrola, C.; Muñoz, F.; Roig-Melo, E.; Panduro, A. The e4 allele of apolipoprotein E gene is a potential risk factor for the severity of macular edema in type 2 diabetic Mexican patients. Ophthalmic Genet. 2002, 23, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Nazará, Z.H.; Ruiz-Madrigal, B.; Martínez-López, E.; Roman, S.; Panduro, A. Association of the epsilon 2 allele of APOE gene to hypertriglyceridemia and to early-onset alcoholic cirrhosis. Alcohol. Clin. Exp. Res. 2008, 32, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, C.A.; Talavera, G.; Ordovas, J.M.; Barriguete, J.A.; Guillen, L.E.; Leco, M.E.; Pedro-Botet, J.; Gonzalez-Barranco, J.; Gomez-Perez, F.J.; Rull, J.A. The apolipoprotein E4 allele is not associated with an abnormal lipid profile in a Native American population following its traditional lifestyle. Atherosclerosis 1999, 142, 409–414. [Google Scholar] [CrossRef]
- Sarkkinen, E.; Korhonen, M.; Erkkilä, A.; Ebeling, T.; Uusitupa, M. Effect of apolipoprotein E polymorphism on serum lipid response to the separate modification of dietary fat and dietary cholesterol. Am. J. Clin. Nutr. 1998, 68, 1215–1222. [Google Scholar] [PubMed]
- Hanson, A.J.; Bayer-Carter, J.L.; Green, P.S.; Montine, T.J.; Wilkinson, C.W.; Baker, L.D.; Watson, G.S.; Bonner, L.M.; Callaghan, M.; Leverenz, J.B.; et al. Effect of apolipoprotein E genotype and diet on apolipoprotein E lipidation and amyloid peptides: Randomized clinical trial. JAMA Neurol. 2013, 70, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Cruz, M.; Tovar, A.R.; Larrieta, E.; Canizales-Quinteros, S.; Torres, N. Increase in HDL-C concentration by a dietary portfolio with soy protein and soluble fiber is associated with the presence of the ABCA1R230C variant in hyperlipidemic Mexican subjects. Mol. Genet. Metab. 2010, 101, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Falchi, M.; El-Sayed Moustafa, J.S.; Takousis, P.; Pesce, F.; Bonnefond, A.; Andersson-Assarsson, J.C.; Sudmant, P.H.; Dorajoo, R.; Al-Shafai, M.N.; Bottolo, L.; et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat. Genet. 2014, 46, 492–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcovecchio, M.L.; Florio, R.; Verginelli, F.; De Lellis, L.; Capelli, C.; Verzilli, D.; Chiarelli, F.; Mohn, A.; Cama, A. Low AMY1 gene copy number is associated with increased body mass index in prepubertal boys. PLoS ONE 2016, 11, e0154961. [Google Scholar] [CrossRef] [PubMed]
- Usher, C.L.; Handsaker, R.E.; Esko, T.; Tuke, M.A.; Weedon, M.N.; Hastie, A.R.; Cao, H.; Moon, J.E.; Kashin, S.; Fuchsberger, C.; et al. Structural forms of the human amylase locus and their relationships to SNPS, haplotypes and obesity. Nat. Genet. 2015, 47, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Jose-Abrego, A.; Panduro, A.; Fierro, N.A.; Roman, S. High prevalence of HBV infection, detection of subgenotypes F1b, A2, and D4, and differential risk factors among Mexican risk populations with low socioeconomic status. J. Med. Virol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gelli, C.; Tarocchi, M.; Abenavoli, L.; Di Renzo, L.; Galli, A.; De Lorenzo, A. Effect of a counseling-supported treatment with the Mediterranean diet and physical activity on the severity of the non-alcoholic fatty liver disease. World J. Gastroenterol. 2017, 23, 3150–3162. [Google Scholar] [CrossRef] [PubMed]
- Abenavoli, L.; Greco, M.; Nazionale, I.; Peta, V.; Milic, N.; Accattato, F.; Foti, D.; Gulletta, E.; Luzza, F. Effects of Mediterranean diet supplemented with silybin-vitamin E-phospholipid complex in overweight patients with non-alcoholic fatty liver disease. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Martínez, P.; Mikhailidis, D.P.; Athyros, V.G.; Bullo, M.; Couture, P.; Covas, M.I.; de Koning, L.; Delgado-Lista, J.; Díaz-López, A.; Drevon, C.A.; et al. Lifestyle recommendations for the prevention and management of metabolic syndrome: An international panel recommendation. Nutr. Rev. 2017, 75, 307–326. [Google Scholar] [CrossRef]
- Roman, S.; Ojeda-Granados, C.; Ramos-Lopez, O.; Panduro, A. Genome-based nutrition: An intervention strategy for the prevention and treatment of obesity and nonalcoholic steatohepatitis. World J. Gastroenterol. 2015, 21, 3449–3461. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Milagro, F.I.; Allayee, H.; Chmurzynska, A.; Choi, M.S.; Curi, R.; De Caterina, R.; Ferguson, L.R.; Goni, L.; Kang, J.X.; et al. Guide for current nutrigenetic, nutrigenomic, and nutriepigenetic approaches for precision nutrition involving the prevention and management of chronic diseases associated with obesity. J. Nutrigenet. Nutrigenom. 2017, 10, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Panduro, A.; Rivera-Iñiguez, I.; Sepulveda-Villegas, M.; Roman, S. Genes, emotions and gut microbiota: The next frontier for the gastroenterologist. World J. Gastroenterol. 2017, 23, 3030–3042. [Google Scholar] [CrossRef] [PubMed]
- Panduro, A.; Ramos-Lopez, O.; Campollo, O.; Zepeda-Carrillo, E.A.; Gonzalez-Aldaco, K.; Torres-Valadez, R.; Roman, S. High frequency of the DRD2/ANKK1 A1 allele in Mexican Native Amerindians and Mestizos and its association with alcohol consumption. Drug Alcohol. Depend. 2017, 172, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Roman, S.; Martinez-Lopez, E.; Fierro, N.A.; Gonzalez-Aldaco, K.; Jose-Abrego, A.; Panduro, A. CD36 genetic variation, fat intake and liver fibrosis in chronic hepatitis C virus infection. World J. Hepatol. 2016, 8, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Panduro, A.; Martinez-Lopez, E.; Roman, S. Sweet Taste Receptor TAS1R2 Polymorphism (Val191Val) Is Associated with a Higher Carbohydrate Intake and Hypertriglyceridemia among the Population of West Mexico. Nutrients 2016, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Roman, S.; Martinez-Lopez, E.; Gonzalez-Aldaco, K.; Ojeda-Granados, C.; Sepulveda-Villegas, M.; Panduro, A. Association of a novel TAS2R38 haplotype with alcohol intake among Mexican-Mestizo population. Ann. Hepatol. 2015, 14, 729–734. [Google Scholar] [PubMed]
- United Nations Educational, Scientfic and Cultural Organization (UNESCO). Available online: https://ich.unesco.org/en/RL/traditional-mexican-cuisine-ancestral-ongoing-community-culture-the-michoacan-paradigm-00400 (assessed on 5 November 2017).
- Ramos, M.A.; Mares, R.E.; Avalos, E.D.; Hernandez, A.; Hernandez, R.; Lameda, R.; Malvaez, A.E.; Rodriguez, C.A.; Rodriguez, R. Pharmacogenetic screening of N-acetyltransferase 2, thiopurine S-methyltransferase, and 5,10-methylene-tetrahydrofolate reductase polymorphisms in Northwestern Nexicans. Genet. Test. Mol. Biomarkers 2011, 15, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Binia, A.; Contreras, A.V.; Canizales-Quinteros, S.; Alonzo, V.A.; Tejero, M.E.; Silva-Zolezzi, I. Geographical and ethnic distribution of single nucleotide polymorphisms within genes of the folate/homocysteine pathway metabolism. Genes Nutr. 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Cubas, C.; Sánchez-Hernández, B.E.; García-Ortiz, H.; Martínez-Hernández, A.; Barajas-Olmos, F.; Cid, M.; Mendoza-Caamal, E.C.; Centeno-Cruz, F.; Ortiz-Cruz, G.; Jiménez-López, J.C.; et al. Heterogenous distribution of MTHFR gene variants among mestizos and diverse Amerindian groups from Mexico. PLoS ONE 2016, 11, e0163248. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Velázquez, R.; Canto, P.; Canto-Cetina, T.; Rangel-Villalobos, H.; Rosas-Vargas, H.; Rodríguez, M.; Canizales-Quinteros, S.; Velázquez Wong, A.C.; Ordoñez-Razo, R.M.; Vilchis-Dorantes, G.; et al. Analysis of polymorphisms in genes (AGT, MTHFR, GPIIIa, and GSTP1) associated with hypertension, thrombophilia and oxidative stress in Mestizo and Amerindian populations of Mexico. Dis. Markers 2010, 28, 323–331. [Google Scholar] [CrossRef]
- Chavez, D.V. Asociación de Ácido Fólico, Homocisteína y Polimorfismo Genético de la Metilentetrahidrofolato-Reductasa con Defectos de Tubo Neural y Labio Hendido Con y Sin Paladar Hendido en Chihuahua. Master’s Thesis, Universidad Autónoma de Nuevo León, Monterrey, Mexico, 2004. [Google Scholar]
- Davalos, I.P.; Olivares, N.; Castillo, M.T.; Cantu, J.M.; Ibarra, B.; Sandoval, L.; Moran, M.C.; Gallegos, M.P.; Chakraborty, R.; Rivas, F. The C677T polymorphism of the methylenetetrahydrofolate reductase gene in Mexican mestizo neural-tube defect parents, control mestizo and native populations. Ann. Genet. 2000, 43, 89–92. [Google Scholar] [CrossRef]
- Rojas, J.C.; Luna, M.; Rangel-Nava, H.; Baños, D.; Collados, M.T. Trombofilia genética y marcadores de activación endotelial en pacientes con preeclampsia. Ginecol. Obstet. Mex. 2010, 78, 401–409. [Google Scholar] [PubMed]
- Aguirre-Rodríguez, A.A.; Martínez, L.E.; Velazco-Campos, M.R.; Sampallo-Hernández, E.; Esmer-Sánchez, M.C. Prevalencia del polimorfismo 677T del gen MTHFR en una muestra de la población de Nuevo León, México. Salud Publica Mex. 2008, 50, 5–7. [Google Scholar]
- León-Cachón, R.B.; Ascacio-Martínez, J.A.; Gamino-Peña, M.E.; Cerda-Flores, R.M.; Meester, I.; Gallardo-Blanco, H.L.; Gómez-Silva, M.; Piñeyro-Garza, E.; Barrera-Saldaña, H.A. A pharmacogenetic pilot study reveals MTHFR, DRD3, and MDR1 polymorphisms as biomarker candidates for slow atorvastatin metabolizers. BMC Cancer 2016, 16, 74. [Google Scholar] [CrossRef] [PubMed]
- Torres-Sánchez, L.; Chen, J.; Díaz-Sánchez, Y.; Palomeque, C.; Bottiglieri, T.; López-Cervantes, M.; López-Carrillo, L. Dietary and genetic determinants of homocysteine levels among Mexican women of reproductive age. Eur. J. Clin. Nutr. 2006, 60, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Guillén Mdel, R.; Torres-Sánchez, L.; Chen, J.; Galván-Portillo, M.; Silva-Zolezzi, I.; Blanco-Muñoz, J.; Hernández-Valero, M.A.; López-Carrillo, L. Dietary consumption of B vitamins, maternal MTHFR polymorphisms and risk of spontaneous abortion. Salud Publica Mex. 2009, 51, 19–25. [Google Scholar] [PubMed]
- Ruiz-Argüelles, G.J.; Coconi-Linares, L.N.; Garces-Eisele, J.; Reyes-Nuñez, V. Methotrexate-induced mucositis in acute leukemia patients is not associated with the MTHFR 677T allele in Mexico. Hematology 2007, 12, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.B.; Lacasaña, M.; Cavazos, R.G.; Borja-Aburto, V.H.; Galaviz-Hernandez, C.; Garduño, C.A. Methylenetetrahydrofolate reductase gene polymorphisms and the risk of anencephaly in Mexico. Mol. Hum. Reprod. 2007, 13, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Antonio-Vejar, V.; Del Moral-Hernández, O.; Alarcón-Romero, L.C.; Flores-Alfaro, E.; Leyva-Vázquez, M.A.; Hernández-Sotelo, D.; Illades-Aguiar, B. Ethnic variation of the C677T and A1298C polymorphisms in the methylenetetrahydrofolate-reductase (MTHFR) gene in southwestern Mexico. Genet. Mol. Res. 2014, 13, 7950–7957. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Herrera, L.; Garcia-Escalante, G.; Castillo-Zapata, I.; Canto-Herrera, J.; Ceballos-Quintal, J.; Pinto-Escalante, D.; Diaz-Rubio, F.; Del Angel, R.M.; Orozco-Orozco, L. Frequency of the thermolabile variant C677T in the MTHFR gene and lack of association with neural tube defects in the State of Yucatan, Mexico. Clin. Genet. 2002, 62, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cuén, J.; Aguilar-Medina, M.; Arámbula-Meraz, E.; Romero-Navarro, J.; Granados, J.; Sicairos-Medina, L.; Ramos-Payán, R. ApoB-100, ApoE and CYP7A1 gene polymorphisms in Mexican patients with cholesterol gallstone disease. World J. Gastroenterol. 2010, 16, 4685–4690. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Fuentes, C.S.; González-Sobrino, B.Z.; Gómez-Sánchez, A.; Martínez Rueda, H.; Chávez-Eakle, R.A.; Serrano Sánchez, C. Distribution of apolipoprotein E alleles in coras and huicholes from Nayarit and Nahuas and mMestizos from Veracruz, Mexico. Hum. Biol. 2005, 77, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, R.; Hernandez-Pacheco, G.; Hesiquio, R.; Zuñiga, J.; Masso, F.; Montaño, L.F.; Ramos-Kuri, M.; Estrada, J.; Granados, J.; Vargas-Alarcon, G. Apolipoprotein E polymorphism in the Indian and Mestizo populations of Mexico. Hum. Biol. 2000, 72, 975–981. [Google Scholar] [PubMed]
- Suástegui, R.A.; Yescas, P.; Guerrero, J.L.; Ochoa, A.; Granados, J.; Jara, A.; López-Caro, O.A.; Alonso, M.E. Frecuencia de la apolipoproteína E en una población nahua. Rev. Investig. Clin. 2002, 54, 415–421. [Google Scholar]
- Kamboh, M.I.; Weiss, K.M.; Ferrell, R.E. Genetic studies of human apolipoproteins. XVI. APOE polymorphism and cholesterol levels in the Mayans of the Yucatan Peninsula, Mexico. Clin. Genet. 1991, 39, 26–32. [Google Scholar] [CrossRef] [PubMed]
| Gene | Function of the Encoded Protein | Polymorphism | Adaptive Allele | Cultural Selection Pressure | Biological Outcome | Population |
|---|---|---|---|---|---|---|
| MTHFR | Folate and homocysteine metabolism | C677T (rs1801133) | T | High intake of folate-rich foods such as leafy greens | A normal metabolism despite a thermolabile enzyme | Amerindians |
| ABCA1 | Interacts with ApoA1 to form nascent HDL | Arg230Cys (rs9282541) | Cys | Maize domestication, low-fat diet | Cholesterol efflux reduction, energy storage | Amerindians |
| APOE | Structural and metabolism-regulatory function of lipoproteins | T388C (rs429358) C526T (rs7412) ε2, ε3, ε4 | ε4 | Low cholesterol and low saturated fat diet | Greater affinity to its receptors, energy storage | Amerindians Sub-Saharans |
| LCT | Lactose digestion | C-13910T (rs4988235) | T | Pastoralism, consumption of milk and derivates | Adult lactase persistence | Europeans |
| AMY1 | Starch hydrolysis | Copy number variation | ≥6 copies | Farming, rich-starch diets | Higher enzyme expression, better starch digestion | Europeans Japanese Hazda hunter-gatherers |
| Genotypes/Alleles | Population | ||||
|---|---|---|---|---|---|
| Ethnic Groups n (%) | Mestizos n (%) | Mestizos-EUR n (%) | p | ||
| MTHFR C677T | Number | 179 a | 952 b | 163 c | |
| CC | 37 (20.7) | 280 (29.4) | 57 (35.0) | ||
| CT+TT | 142 (79.3) | 672 (70.6) | 106 (65.0) | 0.022 * | |
| ABCA1 R230C | Number | 176 a | 540 b | 194 d | |
| RR | 118 (67.0) | 463 (85.7) | 170 (87.6) | ||
| RC+CC | 58 (33.0) | 77 (14.3) | 24 (12.4) | 7.0 × 10−7 * | |
| APOE ε2, ε3, ε4 | Number | 182 a | 637 b | 64 e | |
| ε2 (2n) | 0 (0.0) | 59 (4.6) | 7 (5.5) | ||
| ε3 (2n) | 290 (79.7) | 1094 (85.9) | 117 (91.4) | ||
| ε4 (2n) | 74 (20.3) | 121 (9.5) | 4 (3.1) | 3.0 × 10−8 * | |
| AMY1 | Number | 137 a | 146 b | 148 d | |
| Copy number | 6.77 ± 3.5 | 6.99 ± 3.3 | 6.71 ± 3.1 | ||
| (Χ ± SD) | 0.794 ** | ||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ojeda-Granados, C.; Panduro, A.; Gonzalez-Aldaco, K.; Sepulveda-Villegas, M.; Rivera-Iñiguez, I.; Roman, S. Tailoring Nutritional Advice for Mexicans Based on Prevalence Profiles of Diet-Related Adaptive Gene Polymorphisms. J. Pers. Med. 2017, 7, 16. https://doi.org/10.3390/jpm7040016
Ojeda-Granados C, Panduro A, Gonzalez-Aldaco K, Sepulveda-Villegas M, Rivera-Iñiguez I, Roman S. Tailoring Nutritional Advice for Mexicans Based on Prevalence Profiles of Diet-Related Adaptive Gene Polymorphisms. Journal of Personalized Medicine. 2017; 7(4):16. https://doi.org/10.3390/jpm7040016
Chicago/Turabian StyleOjeda-Granados, Claudia, Arturo Panduro, Karina Gonzalez-Aldaco, Maricruz Sepulveda-Villegas, Ingrid Rivera-Iñiguez, and Sonia Roman. 2017. "Tailoring Nutritional Advice for Mexicans Based on Prevalence Profiles of Diet-Related Adaptive Gene Polymorphisms" Journal of Personalized Medicine 7, no. 4: 16. https://doi.org/10.3390/jpm7040016






