Influence of Amitraz and Oxalic Acid on the Cuticle Proteolytic System of Apis mellifera L. Workers
Abstract
:1. Introduction
2. Results and Discussion
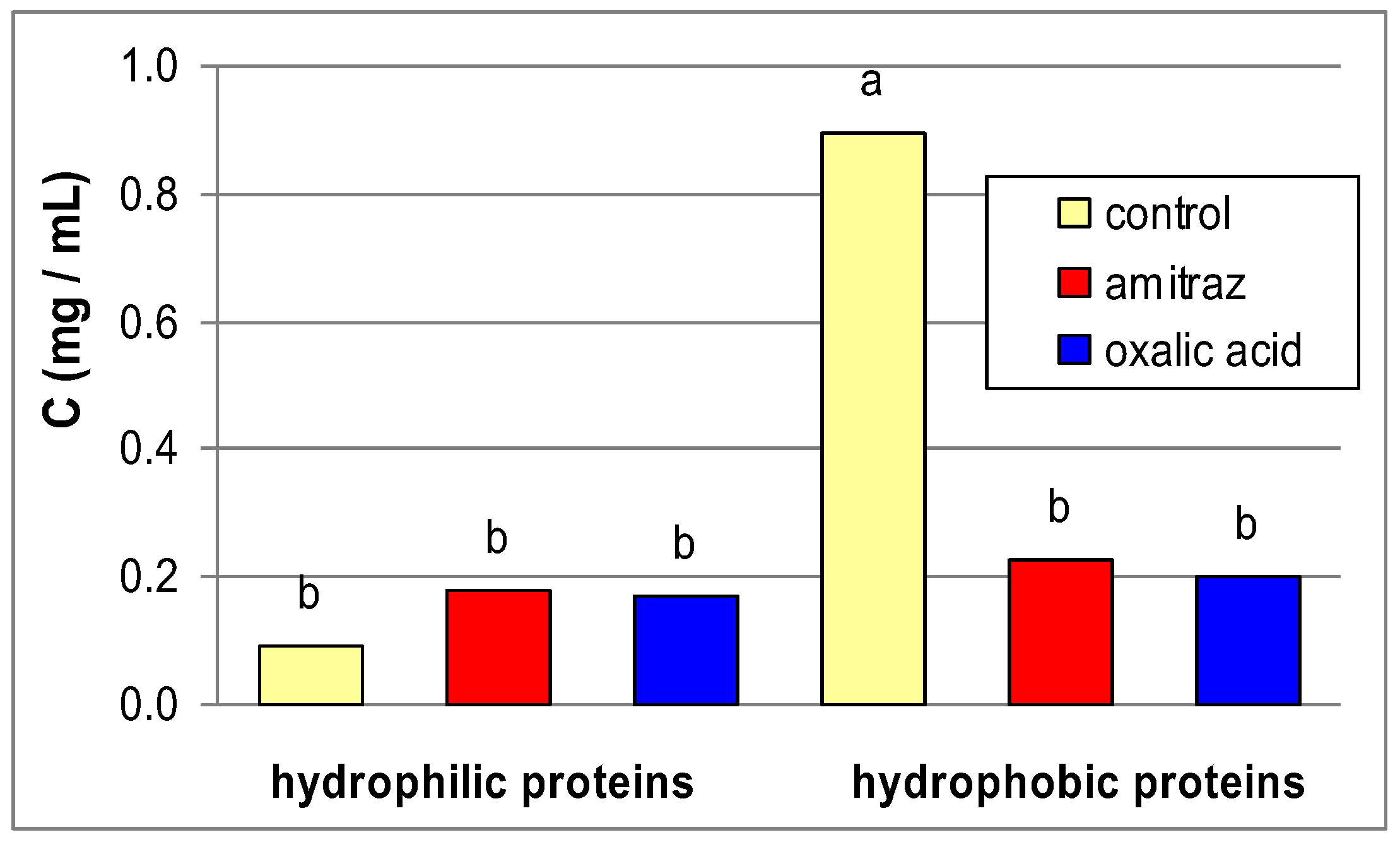
| Control | Amitraz | Oxalic acid | |||
|---|---|---|---|---|---|
| Substrate | proteases | pH | Mean ± se | Mean ± se | Mean ± se |
| Albumin | Hydrophilic (rinsed in water) | 2.4 | 2.17 ± 0.77 | 4.74 ± 3.21 | 2.55 ± 1.79 |
| 7 | 13.88 ± 6.82 | 8.63 ± 3.74 | 7.74 ± 3.42 | ||
| 11.2 | 21.889a ± 5.06 | 20.15a ± 4.61 | 10.52b ± 1.55 | ||
| Hydrophobic (rinsed in triton) | 2.4 | 12.66b ± 0.03 | 13.74b ± 1.93 | 19.57a ± 4.72 | |
| 7 | 7.867 ± 0.05 | 14.53 ± 1.85 | 20.13 ± 5.12 | ||
| 11.2 | 167.71a ± 0.22 | 13.91b ± 1.77 | 19.54c ± 4.70 | ||
| hemoglobin | Hydrophilic (rinsed in water) | 2.4 | 1.87 ± 0.87 | 1.00 ± 0.50 | 1.00 ± 1.00 |
| 7 | 1.73 ± 0.16 | 1.08 ± 0.43 | 1.79 ± 1.54 | ||
| 11.2 | 2.56a ± 0.33 | 0.44b ± 0.16 | 1.55ab ± 0.83 | ||
| Hydrophobic(rinsed in triton) | 2.4 | 15.58a ± 0.14 | 5.67b ± 0.99 | 2.87c ± 0.71 | |
| 7 | 20.64a ± 0.07 | 5.22b ± 1.15 | 4.38b ± 1.17 | ||
| 11.2 | 10.72a ± 0.07 | 2.96b ± 1.23 | 3.29b ± 0.72 | ||
| cytochrome C | Hydrophilic (rinsed in water) | 2.4 | 4.65a ± 0.61 | 0.15b ± 0.07 | 1.53c ± 0.15 |
| 7 | 7.42a ± 0.82 | 0.93b ± 0.11 | 0.68b ± 0.25 | ||
| 11.2 | 9.54a ± 0.31 | 2.19b ± 0.23 | 1.05c ± 0.11 | ||
| Hydrophobic (rinsed in triton) | 2.4 | 12.27a ± 0.10 | 8.92b ± 1.15 | 12.33a ± 1.77 | |
| 7 | 6.03b ± 0.19 | 6.57a ± 0.57 | 6.71a ± 0.21 | ||
| 11.2 | 22.13a ± 0.16 | 6.66b ± 0.44 | 3.39c ± 0.26 | ||
| ovoalbumin | Hydrophilic (rinsed in water) | 2.4 | 1.85a ± 0.16 | 0.51b ± 0.09 | 0.65c ± 0.22 |
| 7 | 14.54a ± 0.08 | 1.86b ± 0.36 | 0.00c ± 0.00 | ||
| 11.2 | 4.18a ± 0.21 | 0.00b ± 0.00 | 0.69b ± 0.17 | ||
| Hydrophobic (rinsed in triton) | 2.4 | 51.18a ± 0.30 | 4.17b ± 0.31 | 2.72c ± 0.43 | |
| 7 | 23.71a ± 0.09 | 3.00b ± 0.49 | 2.178c ± 0.36 | ||
| 11.2 | 48.32a ± 0.22 | 3.02b ± 0.56 | 3.35b ± 0.53 | ||
2.1. Cuticle Proteases
| Control—no treatment | Amitraz treatment | Oxalic acid treatment | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hydrophilic protein | hydrophobic protein | hydrophilic protein | hydrophobic protein | hydrophilic protein | hydrophobic protein | ||||||||
| pH | Rf (mm) | Band number | OD | Band number | OD | Band number | OD | Band number | OD | Band number | OD | Band number | OD |
| 2.4 | 0–40 | 1 | 0.25 | 3 | 0.86 | 3 | 0.21 | 4 | 0.25 | 3 | 0.15 | 1 | 0.20 |
| 41–60 | 1 | 0.40 | 2 | 0.35 | 1 | 0.26 | 3 | 0.43 | 1 | 0.35 | 1 | 0.23 | |
| 61–100 | 4 | 0.28 | 0 | 0 | 3 | 0.36 | 4 | 0.46 | 1 | 0.49 | 3 | 0.53 | |
| 7.0 | 0–40 | 5 | 0.18 | 2 | 0.54 | 1 | 0.49 | 4 | 0.66 | 0 | 0 | 3 | 0.48 |
| 41–60 | 2 | 0.20 | 0 | 0 | 1 | 0.63 | 1 | 0.92 | 2 | 0.47 | 1 | 0.74 | |
| 61–100 | 4 | 0.25 | 3 | 0.36 | 2 | 0.61 | 2 | 0.68 | 3 | 0.52 | 2 | 0.74 | |
| 11.0 | 0–40 | 3 | 0.25 | 0 | 0 | 1 | 0.30 | 5 | 0.23 | 1 | 0.12 | 2 | 0.14 |
| 41–60 | 0 | 0 | 0 | 0 | 1 | 0.23 | 1 | 0.15 | 1 | 0.19 | 1 | 0.20 | |
| 61–100 | 1 | 0.32 | 0 | 0 | 4 | 0.25 | 1 | 0.18 | 2 | 0.18 | 3 | 0.38 | |
2.2. Cuticle Protease Inhibitors
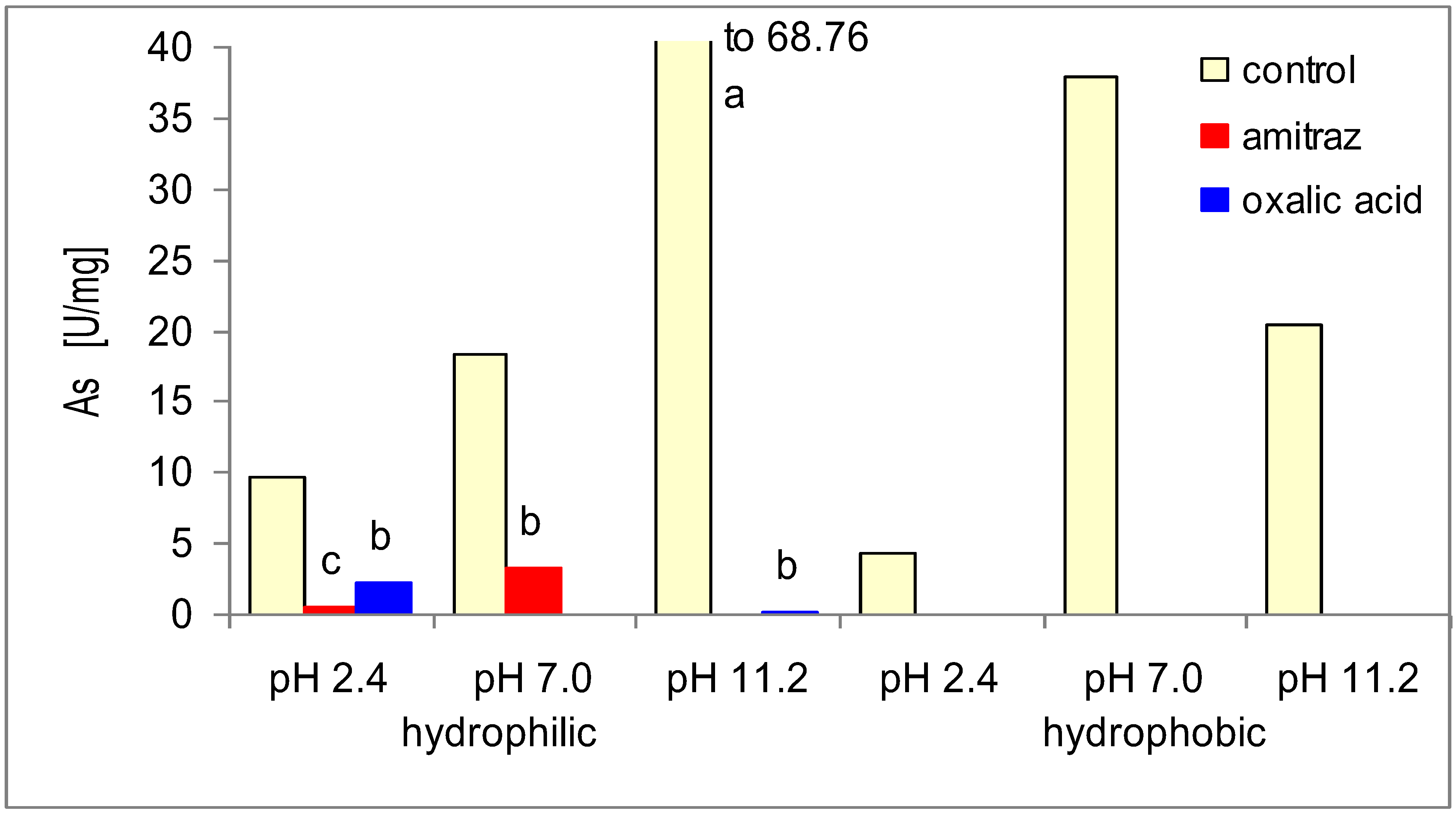

| Control—no treatment | Amitraz treatment | Oxalic acid treatment | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hydrophilic protein | hydrophobic protein | hydrophilic protein | hydrophobic protein | hydrophilic protein | hydrophobic protein | ||||||||
| pH | Rf (mm) | Band number | OD | Band number | OD | Band number | OD | Band number | OD | Band number | OD | Band number | OD |
| 2.4 | 0–40 | 4 | 0.21 | 0 | 0 | 5 | 0.73 | 2 | 0.74 | 4 | 0.65 | 5 | 0.63 |
| 41–60 | 0 | 0 | 1 | 0.20 | 1 | 0.58 | 1 | 0.67 | 1 | 0.55 | 0 | 0 | |
| 61–100 | 3 | 0.21 | 4 | 0.14 | 5 | 0.53 | 4 | 0.56 | 2 | 0.58 | 3 | 0.61 | |
| 7.0 | 0–40 | 2 | 0.16 | 1 | 0.23 | 2 | 0.23 | 4 | 0.35 | 3 | 0.26 | 1 | 0.23 |
| 41–60 | 0 | 0 | 0 | 0 | 1 | 0.30 | 2 | 0.52 | 0 | 0 | 0 | 0 | |
| 61–100 | 0 | 0 | 1 | 0.21 | 1 | 0.27 | 4 | 0.34 | 2 | 0.26 | 5 | 0.39 | |
| 11.0 | 0–40 | 3 | 0.37 | 4 | 0.23 | 2 | 0.14 | 2 | 0.16 | 2 | 0.08 | 0 | 0 |
| 41–60 | 0 | 0 | 0 | 0 | 2 | 0.10 | 0 | 0 | 1 | 0.11 | 0 | 0 | |
| 61–100 | 3 | 0.36 | 0 | 0 | 0 | 0 | 3 | 0.28 | 1 | 0.04 | 3 | 0.23 | |
2.3. Apian Cuticle Proteolytic System — General Comments
2.4. Activity towards Model Microorganisms
| Group | Antifungal activity | Antibacterial activity | ||||
|---|---|---|---|---|---|---|
| Candida albicans | Aspergillus niger | Bacillus subtilis | Staphylococcus aureus | Salmonella typhimurium | Pseudomonas aeruginosa | |
| Control | 978.57 | 23.36 | 78.36 | 308.61 | 126.49 | 101.33 |
| Amitraz | 212.50 | 0 | 756.51 | 0 | 259.62 | 207.24 |
| Oxalic acid | 340.69 | 0 | 154.18 | 0 | 0 | 0 |
3. Experimental Section
3.1. The Experimental Design
3.2. Sampling and the Data Base
- 2 mL—for determining protease and protease inhibitor activities (portion a),
- 2 mL—for electrophoretic assays (portion b),
- 2 mL—for determining antifungal and antibacterial activities in vivo (portion c),
- 2 mL—reserve (portion d).
3.3. The Analytical Procedures
- the general protein content by the Lowry method, as modified by Schacterle and Pollack [31];
- the proteolytic activity in relation to the diagnostic inhibitors of proteolytic enzymes (pepstatin A, PMSF, iodoacetamide, o-phenanthroline), by the Lee and Lin method [33];
- the activities of acidic, neutral, and alkaline proteases according to the Anson method [32];
- the levels of natural inhibitors of acidic, neutral, and alkaline proteases, based on the Lee and Lin method [33].
3.4. In Vivo Laboratory Tests
- SABG [36]—to determine activity in relation to Aspergillus niger,
- YPD [37]—to determine activity in relation to Candida albicans,
- LB [38]—to determine activity in relation to Staphylococcus ureus (ATCC 25923), Bacillus subtilis (ATCC 6633), Micrococcus luteus (ATCC 7468), Salmonella typhimurium (ATCC 13311), Pseudomonas aeruginosa (ATCC 17853), and Escherichia coli (ATCC 10536).
3.5. Statistics
4. Conclusion
References and Notes
- Terra, W.R.; Ferreira, C. Insect digestive enzymes: Properties, compartmentalization and function. Comp. Biochem. Physiol. 1994, 109, 1–62. [Google Scholar] [CrossRef]
- Barrett, A.J. Peptidases: A view of classification and nomenclature. In Proteases: New Perspectives, 2nd ed.; Turk, V., Ed.; Birkhãuser: Berlin, German, 1999; Volume 1, pp. 1–12. [Google Scholar]
- Bode, W.; Fernandez-Catalan, C.; Nagase, H.; Maskos, K. Endoproteinase—Protein inhibitor interaction. APMIS 1999, 107, 3–10. [Google Scholar] [CrossRef]
- Otlewski, J.; Jaskólski, M.; Buczek, O.; Cierpiński, T.; Czapińska, H.; Krowarsch, D.; Smalas, A.O.; Stachowiak, D.; Szpineta, A.; Dadlez, M. Structure—Function relationship of serine protease—Protein inhibitor interaction. Acta Bioch. Pol. 2001, 48, 419–428. [Google Scholar]
- Deraison, M.C. Isolement, caractérisation et cibles de nouveaux inhibiteurs de protéases pour la création de plantes transgéniques résistantes aux pucerons (in French). Universite Paris XI UFR Scientique d’orsay 2004, 1, 1–241. [Google Scholar]
- Lima, P.R.M.; Brochetto-Braga, M.R.; Chaud-Netto, J. Proteolytic activity of Africanized honeybee (Apis mellifera: hymenoptera, apidae) venom. J. Venom. Anim. Toxins 2000, 6, 104–113. [Google Scholar]
- Sagili, R.R.; Panjiew, T.; Zhu-Salzman, K. Effects of soybean trypsin inhibitor on hypopharyngeal gland protein content, total midgut protease activity and survival of the honey bee (Apis mellifera L.). J. Insect. Physiol. 2005, 51, 953–995. [Google Scholar] [CrossRef]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bee Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef]
- Grzywnowicz, K.; Ciołek, A.; Tabor, A.; Jaszek, M. Profiles of body-surface proteolytic system of honey bee: Proteases and their natural inhibitors during ontogenesis and seasonal changes of Apis mellifera casts. Apidolgie 2009, 40, 4–19. [Google Scholar] [CrossRef]
- Strachecka, A.; Paleolog, J.; Grzywnowicz, K. The surface proteolytic activity in Apis mellifera. J. Apic. Sci. 2008, 52, 49–56. [Google Scholar]
- Strachecka, A.; Gryzińska, M.; Krauze, M. The influence of environmental pollution on the protective proteolytic barrier of the honey bee Apis mellifera’s body surface. Pol. J. Environ. Stud. 2010, 19, 855–859. [Google Scholar]
- Cummins, J. Requiem for the honeybee. Sci. Soc. 2007, 34, 37–38. [Google Scholar]
- Ho, M.W.; Cummins, J. Mystery of disappearing honeybees. Sci. Soc. 2007, 34, 35–36. [Google Scholar]
- Blanchard, P.; Schurr, F.; Celle, O.; Cougoule, N.; Drajnudel, P.; Thiery, R.; Faucon, J.P.; Ribière, M. First detection of Israeli acute paralysis virus (IAPV) in France, a dicistrovirus affecting honeybees (Apis mellifera). J. Invertebr. Pathol. 2008, 99, 348–350. [Google Scholar] [CrossRef]
- Kevan, P.G.; Guzman, E.; Skinder, A.; van Englesdorp, D. Colony collapse disorder (CCD) in Canada: Do we have problem? Amer. Bee J. 2005, 145, 507–509. [Google Scholar]
- Johnson, R. Recent Honey Bee Colony Declines; Report for US Congress Senate Committee on Agriculture; Congressional Research Service, The Library of Congress: Washington, DC, USA, 20 June 2007; pp. 1–9. [Google Scholar]
- Rosenkranz, P.; Aumeier, P.; Zigelmann, B. Biology and control of Varroa destructor. J. Invertebr Pathol. 2010, 103, S96–S119. [Google Scholar] [CrossRef]
- Milani, N. The resistance of Varroa jacobsoni Oud. to pyrethroids a laboratory assay. Apidologie 1995, 26, 415–429. [Google Scholar] [CrossRef]
- Elzen, P.J.; Baxter, J.R.; Spivak, M.; Wilson, W.T. Control of Varroa jacobsoni Oud. Resistant to fluvalinate and amitraz using coumaphos. Apidologie 2000, 31, 437–441. [Google Scholar] [CrossRef]
- Lipiński, Z.; Szubstarski, J. Resistance of Varroa destructor to most commonly used synthetic acaricides. Pol. J. Vet. Sci. 2007, 10, 289–294. [Google Scholar]
- Higes, M.; Meana, A.; Suarez, M.; Llorente, J. Negative long-term effects on bee colonies treated with oxalic acid against Varroa jacobsoni Oud. Apidologie 1999, 30, 289–292. [Google Scholar] [CrossRef]
- Imdorf, A.; Charrière, J.D.; Kilchenmann, V.; Bogdanov, S.; Fluri, P. Alternative strategy in central Europe for the control of Varroa desrtuctor in honey bee colonies. Apiacta 2003, 38, 258–285. [Google Scholar]
- Howis, M.; Chorbiński, P.; Nowakowski, P. Influence of exposure to formic acid on the physiological status of the apian (Apis mellifera L.) midgut. In Proceedings of the XLVIII Conference, Scientific Apiculture Conference, Puławy, Poland, 5–7 April 2010; ISiK Press: Puławy, Poland, 2010; p. 21. [Google Scholar]
- Rademacher, E.; Harz, M. Oxalic acid for the control of varroosis in honey bee colonies—A review. Apidologie 2006, 37, 98–120. [Google Scholar] [CrossRef]
- Chuda-Mickiewicz, B.; Prabucki, J.; Kazimierczak, J.; Samborsk, J. The varroacidal efficacy of Amitraz. Med. Wet. 2007, 63, 110–112. [Google Scholar]
- Paleolog, J. Direct mutual contacts with beekeepers during their annual meetings and training workshops, Poland, 2010. President of the Polish Bee Research Association, Warsaw, Mazovian Voivodship, Poland.
- Loucif-Ayad, W.; Aribi, N.; Smagghe, G.; Soltani, N. A scientific note on the impact of acaracides on the nutritional biochemistry of Apis mellifera intermissa (Hymenoptera: Apidae). Apidologie 2010, 41, 135–137. [Google Scholar] [CrossRef]
- Paleolog, J.; Strachecka, A.; Burzyński, S.R.; Olszewski, K.; Borsuk, G. The larval diet supplemented with sodium phenylacetylglutaminate influences the worker cuticle proteolytic system in western honey bee Apis mellifera L. J. Apic. Sci. 2011, 55, 73–84. [Google Scholar]
- Currie, C.R.A. Community of ants, fungi and bacteria: A multilateral approach to studying symbiosis. Ann. Rev. Microbiol. 2001, 55, 357–380. [Google Scholar] [CrossRef]
- Frączek, R.; Żółtowska, K.; Lipiński, Z.; Dymitriuk, M. Proteolytic activity in the extracts and in the excretory/secretory products from Varroa destructor parasitic mite of honeybee. Int. J. Acarol. 2012, 38, 101–109. [Google Scholar] [CrossRef]
- Schacterle, G.; Pollack, R. Simplified method for quantitative assay of small amounts of protein in biological material. Anal. Biochem 1973, 51, 654–655. [Google Scholar]
- Anson, M. The estimation of pepsin, tripsin, papain and cathepsin with hemoglobin. J. Gen. Physiol. 1938, 22, 79–84. [Google Scholar] [CrossRef]
- Lee, T.; Lin, Y. Trypsin inhibitor and trypsin—Like protease activity in air—Or submergence—Grown rice (Oryza sativa L.) coleoptiles. Plant Sci. 1995, 106, 43–54. [Google Scholar] [CrossRef]
- Laemmli, U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Felicioli, R.; Garzelli, B.; Vaccari, L.; Melfi, D.; Balestreri, E. Activity staining of protein inhibitors of proteases on gelatin-containing polyacrylamide gel electrophoresis. Anal. Biochem. 1997, 244, 176–179. [Google Scholar]
- Sabouraud, R. Contribution a l'etude de la trichophytie humaine. Etude clinique, microscopique et bacterioloqique sur la pluralite des trichophytons de l'homme (French). Ann. Dermatol. Syphil. 1892, 3, 1061–1087. [Google Scholar]
- Murthy, M.S.; Rao, B.S.; Reddy, N.M.; Subrahmanyam, P.; Madhvanath, U. Non-equivalence of YEPD and synthetic complete media in yeast reversion studies. Mutat. Res. 1975, 2, 219–223. [Google Scholar]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1952, 3, 293–300. [Google Scholar]
- MultiScanBase, Computer Scanning Systems II: Warsaw, Poland, 1993/2004; Computer Scanning Systems, CSS Scan; license no: 148.236.318.
- SAS, SAS Institute: Cary, NC, USA, 2002–2003; version 9.1, revision 9.1.3; license 86636, order 539426.
- Strachecka, A.; Paleolog, J.; Borsuk, G.; Olszewski, K. The influence of formic acid on the body surface proteolytic system at different developmental stages in Apis mellifera L. workers. J. Apic. Res. 2012, 51, 252–262. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Strachecka, A.; Paleolog, J.; Olszewski, K.; Borsuk, G. Influence of Amitraz and Oxalic Acid on the Cuticle Proteolytic System of Apis mellifera L. Workers. Insects 2012, 3, 821-832. https://doi.org/10.3390/insects3030821
Strachecka A, Paleolog J, Olszewski K, Borsuk G. Influence of Amitraz and Oxalic Acid on the Cuticle Proteolytic System of Apis mellifera L. Workers. Insects. 2012; 3(3):821-832. https://doi.org/10.3390/insects3030821
Chicago/Turabian StyleStrachecka, Aneta, Jerzy Paleolog, Krzysztof Olszewski, and Grzegorz Borsuk. 2012. "Influence of Amitraz and Oxalic Acid on the Cuticle Proteolytic System of Apis mellifera L. Workers" Insects 3, no. 3: 821-832. https://doi.org/10.3390/insects3030821





