Individual Variability of Nosema ceranae Infections in Apis mellifera Colonies
Abstract
:1. Introduction
2. Results and Discussion
| Spring | Summer | Fall | ||||
|---|---|---|---|---|---|---|
| Colony | Avg. Copy # 1 | N 2 | Avg. Copy # | N | Avg. Copy # | N |
| 1 | 1099.66 ± 1099.66 | 1/43 | 159871.87 ± 99440.99 | 19/50 | 0 ± 0 | 0/50 |
| 2 | 0.61 ± 0.61 | 1/50 | 6.52 ± 1.62 | 16/50 | 0 ± 0 | 0/50 |
| 3 | 42400.72 ± 28832.82 | 15/39 | 6.05 ± 1.67 | 13/50 | 1.41 ± 1.41 | 1/50 |
| 4 | 250.45 ± 250.45 | 1/44 | 0 ± 0 | 0/43 | 0 ± 0 | 0/50 |
| 5 | 773.69 ± 450.84 | 3/36 | 20212.60 ± 14470.56 | 6/50 | 0 ± 0 | 0/50 |
| Spring | Summer | Fall | ||||
|---|---|---|---|---|---|---|
| Colony | Avg. # Spores 1 | N 2 | Avg. # Spores | N | Avg. # Spores | N |
| 1 | 5988 ± 5929 | 3/43 | 19950 ± 11919 | 7/50 | 0 ± 0 | 0/50 |
| 2 | 50 ± 35 | 2/50 | 75 ± 55 | 2/50 | 0 ± 0 | 0/50 |
| 3 | 68622 ± 19132 | 14/39 | 50 ± 35 | 2/50 | 25 ± 25 | 1/50 |
| 4 | 85 ± 63 | 2/44 | 145 ± 62 | 5/43 | 25 ± 25 | 1/50 |
| 5 | 27500 ± 16044 | 3/36 | 148525 ± 98311 | 7/50 | 50 ± 35 | 2/50 |
| Spring | Summer 3 | Fall | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Colony | Spore Counts | Raw qPCR 1 | Threshold qPCR 2 | Spore Counts | Raw qPCR | Threshold qPCR | Spore Counts | Raw qPCR | Threshold qPCR |
| 1 | 7 | 32.6 | 2.3 | 14 | 82 | 38 | 0 | 12 | 0 |
| 2 | 4 | 36 | 2 | 4 | 56 | 32 | 0 | 2 | 0 |
| 3 | 35.9 | 84.6 | 38.5 | 4 | 56 | 26 | 2 | 14 | 2 |
| 4 | 4.5 | 63.6 | 2.3 | 11.6 | 16.3 | 0 | 2 | 10 | 0 |
| 5 | 3 | 63.9 | 8.3 | 14 | 18 | 12 | 4 | 12 | 0 |
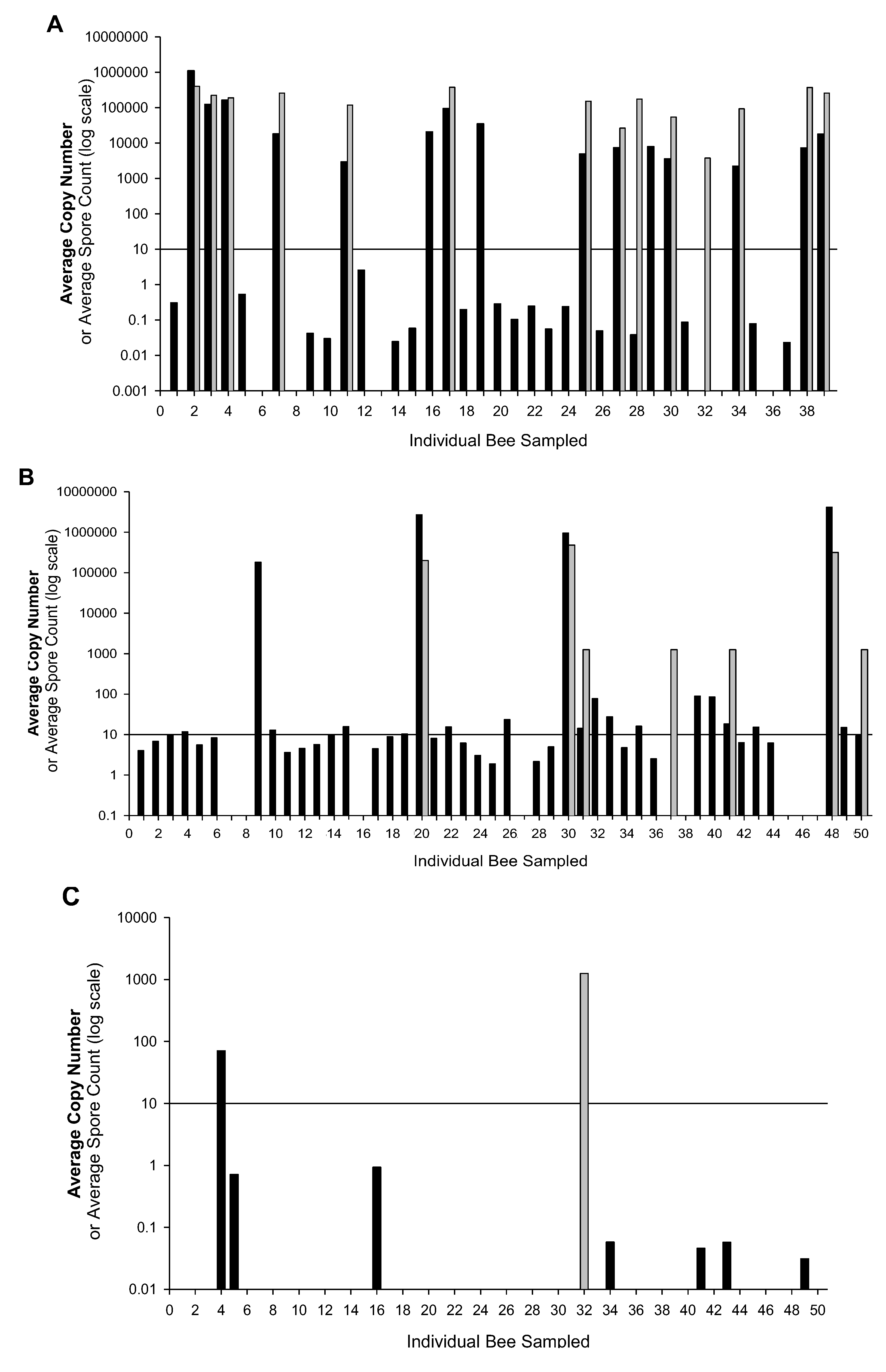
3. Experimental Section
3.1. Sample Collection
3.2. Genomic DNA Extraction
3.3. Spore Counting
3.4. Quantitative Real-time PCR (qPCR)
3.5. Statistical Analysis
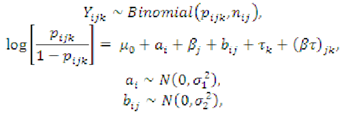
4. Conclusions
Acknowledgments
References and Notes
- Fries, I. Protozoa. In Honey Bee Pests, Predators, & Diseases, 3rd ed.; Morse, R.A., Flottum, K., Eds.; A.I. Root Company: Medina, Saudi Arabia, 1997; pp. 59–76. [Google Scholar]
- Bailey, L. The measurement and interrelationships of infections with Nosema apis and Malpighamoeba mellificae of honey-bee populations. J. Invertebr. Pathol. 1968, 12, 175–179. [Google Scholar] [CrossRef]
- Bailey, L. Infectious Diseases of the Honey Bee, 1st ed.; Land Books: London, 1963. [Google Scholar]
- Fries, I.; Feng, F.; da Silva, A.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Fries, I.; Martín, R.; Meana, A.; García-Palencia, P.; Higes, M. Natural infections of Nosema ceranae in European honey bees. J. Apic. Res. 2006, 45, 230–233. [Google Scholar]
- Higes, M.; Martín, R.; Meana, A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 2006, 92, 93–95. [Google Scholar] [CrossRef]
- Huang, W.-F.; Jiang, J.-H.; Chen, Y.-W.; Wang, C.-H. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie 2007, 38, 30–37. [Google Scholar] [CrossRef]
- Dyess, E. A study of the seasonal variations of Nosema apis Zander of honey bees in Mississippi. Am. Bee J. 1978, 118, 33–35. [Google Scholar]
- Fries, I. Nosema apis- A parasite in the honey bee colony. Bee World 1993, 74, 5–19. [Google Scholar]
- Farrar, C.L. Nosema disease contributes to winter losses and queen supersedure. Gleanings Bee Culture 1942, 70, 660-661, 701. [Google Scholar]
- Moeller, F.E. Nosema disease- its control in honey bee colonies. In Technical Bulletin No. 1569; USDA; Science and Education Administration: Washington, DC, USA, 1978. [Google Scholar]
- Jeffree, E.P.; Allen, M.D. The influence of colony size and of nosema disease on the rate of population loss in honey bee colonies in winter. J. Econ. Entomol. 1956, 49, 831–834. [Google Scholar]
- Higes, M.; Martín-Hernández, R.; Botías, C.; Bailón, E.G.; González-Porto, A.V.; Barrios, L.; Nozal, M.J.d.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G.; Meana, A. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailón, E.; González-Porto, A.; García-Palencia, P.; Meana, A.; María; Mayo, R.; Bernal, J. Honeybee colony collapse due to Nosema ceranae in professional apiaries. Environ. Microbiol. Reports 2009, 1, 110–113. [Google Scholar] [CrossRef]
- Traver, B.E.; Fell, R.D. Prevalence and infection intensity of Nosema in honey bee (Apis mellifera L.) colonies in Virginia. J. Invertebr. Pathol. 2011, 107, 43–49. [Google Scholar] [CrossRef]
- Mussen, E.C.; Furgala, B.; Hyser, R.A. Enzootic levels of Nosema disease in the continental United States (1074). Am. Bee J. 1975, 115, 48-50, 58. [Google Scholar]
- Fries, I. Nosema ceranae in European honey bees (Apis mellifera). J. Invertebr. Pathol. 2010, 103, S73–S79. [Google Scholar] [CrossRef]
- Jaycox, E.R. Surveys for Nosema disease in honey bees in California. J. Econ. Entomol. 1960, 53, 95–98. [Google Scholar]
- Doull, K.M.; Eckert, J.E. A survey of the incidence of Nosema disease in California. J. Econ. Entomol. 1962, 55, 313–317. [Google Scholar]
- Doull, K.M.; Cellier, K.M. A survey of the incidence of nosema disease (Nosema apis Zander) of the honey bee in South Australia. J. Insect Pathol. 1961, 3, 280–288. [Google Scholar]
- Pickard, R.S.; El-Shemy, A.A.M. Seasonal variation in the infection of honeybee colonies with Nosema apis Zander. J. Apic. Res. 1989, 28, 93–100. [Google Scholar]
- Martin-Hernandez, R.; Meana, A.; Prieto, L.; Salvador, A.M.; Garrido-Bailon, E.; Higes, M. Outcome of the colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007, 73, 6331–6338. [Google Scholar] [CrossRef]
- Traver, B.E.; Williams, M.R.; Fell, R.D. Comparison of within hive sampling and seasonal activity of Nosema ceranae in honey bee colonies. J. Invertebr. Pathol. 2012, 109, 187–193. [Google Scholar] [CrossRef]
- Bailey, L. The natural mechanism of suppression of Nosema apis Zander in enzootically infected colonies of the honey bee, Apis mellifera Linnaeus. J. Insect Pathol. 1959, 1, 347–350. [Google Scholar]
- Higes, M.; Martín-Hernández, R.; Meana, A. Nosema ceranae in Europe: an emergent type C nosemosis. Apidologie 2010, 41, 375–392. [Google Scholar] [CrossRef]
- Gisder, S.; Hedtke, K.; Mockel, N.; Frielitz, M.C.; Linde, A.; Genersch, E. Five-Year Cohort Study of Nosema spp. in Germany: Does Climate Shape Virulence and Assertiveness of Nosema ceranae? Appl. Environ. Microbiol. 2010, 76, 3032–3038. [Google Scholar] [CrossRef]
- Copley, T.R.; Jabaji, S.H. Honeybee glands as possible infection reservoirs of Nosema ceranae and Nosema apis in naturally infected forager bees. J. Appl. Microbiol. 2012, 112, 15–24. [Google Scholar] [CrossRef]
- Martin-Hernandez, R.; Meana, A.; Garcia-Palencia, P.; Marin, P.; Botias, C.; Garrido-Bailon, E.; Barrios, L.; Higes, M. Effect of temperature on the biotic potential of honeybee microsporidia. Appl. Environ. Microbiol. 2009, 75, 2554–2557. [Google Scholar]
- Meana, A.; Martín-Hernández, R.; Higes, M. The reliability of spore counts to diagnose Nosema ceranae infections in honey bees. J. Apic. Res. 2010, 49, 212–214. [Google Scholar] [CrossRef]
- Cantwell, G.E. Standard methods for counting Nosema spores. Am. Bee J. 1970, 110, 22–223. [Google Scholar]
- OIE Chapter 2.2.4 Nosemosis of honey bees. Available online: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.02.04_NOSEMOSIS.pdf (acessed on 14 October 2012).
- L'Arrivee, J.C.M. The effects of sampling sites on Nosema determination. J. Insect Pathol. 1963, 5, 349–355. [Google Scholar]
- Seeley, T.D. The Wisdom of the Hive: The Social Physiology of Honey Bee colonies; Harvard University Press: Cambridge, Massachusetts, UK, 1995; p. 295. [Google Scholar]
- Fries, I.; Ekbohm, G.; Villumstad, E. Nosema apis, sampling techniques and honey yield. J. Apic. Res. 1984, 23, 102–105. [Google Scholar]
- Paxton, R.J.; Klee, J.; Korpela, S.; Fries, I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie 2007, 38, 558–565. [Google Scholar] [CrossRef]
- Wu, J.Y.; Smart, M.D.; Anelli, C.M.; Sheppard, W.S. Honey bees (Apis mellifera) reared in brood combs containing high levels of pesticide residues exhibit increased susceptibility to Nosema (Microsporidia) infection. J. Invertebr. Pathol. 2012, 109, 326–329. [Google Scholar] [CrossRef]
- Muz, M.N.; Girisgin, A.O.; Muz, D.; Aydin, L. Molecular detection of Nosema ceranae and Nosema apis infections in Turkish apiaries with collapsed colonies. J. Apic. Res. 2010, 49, 342. [Google Scholar] [CrossRef]
- Suwannapong, G.; Yemor, T.; Boonpakdee, C.; Benbow, M.E. Nosema ceranae, a new parasite in Thai honeybees. J. Invertebr. Pathol. 2011, 106, 236–241. [Google Scholar] [CrossRef]
- Chaimanee, V.; Warrit, N.; Chantawannakul, P. Infections of Nosema ceranae in four different honeybee species. J. Invertebr. Pathol. 2010, 105, 207–210. [Google Scholar] [CrossRef]
- Yoshiyama, M.; Kimura, K. Distribution of Nosema ceranae in the European honeybee, Apis mellifera in Japan. J. Invertebr. Pathol. 2011, 106, 263–267. [Google Scholar] [CrossRef]
- Giersch, T.; Berg, T.; Galea, F.; Hornitzky, M. Nosema ceranae infects honey bees (Apis mellifera) and contaminates honey in Australia. Apidologie 2009, 40, 117–123. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailón, E.; Botías, C.; Meana, A. The presence of Nosema ceranae (Microsporidia) in North African honey bees (Apis mellifera intermissa). J. Apic. Res. 2009, 48, 217–219. [Google Scholar] [CrossRef]
- Invernizzi, C.; Abud, C.; Tomasco, I.H.; Harriet, J.; Ramallo, G.; Campá, J.; Katz, H.; Gardiol, G.; Mendoza, Y. Presence of Nosema ceranae in honeybees (Apis mellifera) in Uruguay. J. Invertebr. Pathol. 2009, 101, 150–153. [Google Scholar] [CrossRef]
- Calderón, R.A.; Sanchez, L.A.; Yañez, O.; Fallas, N. Presence of Nosema ceranae in africanized honey bee colonies in Costa Rica. J. Apic. Res. 2008, 47, 328–329. [Google Scholar]
- Williams, G.R.; Shafer, A.B.A.; Rogers, R.E.L.; Shutler, D.; Stewart, D.T. First detection of Nosema ceranae, a microsporidian parasite of European honey bees (Apis mellifera), in Canada and central USA. J. Invertebr. Pathol. 2008, 97, 189–192. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.D.; Smith, I.B.; Pettis, J.S. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J. Invertebr. Pathol. 2008, 97, 186–188. [Google Scholar] [CrossRef]
- Topolska, G.; Kasprzak, S. First cases of Nosema ceranae infection in bees in Poland. Medycyna Weterynaryjna 2007, 63, 1504–1506. [Google Scholar]
- Klee, J.; Besana, A.M.; Genersch, E.; Gisder, S.; Nanetti, A.; Tam, D.Q.; Chinh, T.X.; Puerta, F.; Ruz, J.M.; Kryger, P.; Message, D.; Hatjina, F.; Korpela, S.; Fries, I.; Paxton, R.J. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 2007, 96, 1–10. [Google Scholar] [CrossRef]
- Paxton, R.J.; Klee, J.; Korpela, S.; Fries, I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie 2007, 38, 558–565. [Google Scholar] [CrossRef]
- Chauzat, M.P.; Higes, M.; Martin-Hernandez, R.; Meana, A.; Cougoule, N.; Faucon, J.P. Presence of Nosema ceranae in French honey bee colonies. J. Apic. Res. 2007, 46, 127–128. [Google Scholar]
- Farrar, C.L. Fumagillin for nosema control in package bees. Am. Bee J. 1954, 94, 52-53, 60. [Google Scholar]
- Gochnauer, T.A. Fumidil B and Nosema control in wintering colonies. Glean. Bee Cult. 1957, 85, 528–529. [Google Scholar]
- Doull, K.M. The effects of time of day and method of sampling on the determination of nosema disease in beehives. J. Invertebr. Pathol. 1965, 7, 1–4. [Google Scholar] [CrossRef]
- L'Arrivee, J.C.M. Comparison of composite versus individual bee sampling for Nosema apis Zander. Journal of Insect Pathology 1963, 5, 434–439. [Google Scholar]
- Bailey, L. The transmission of nosema disease. Bee World 1953, 34, 171–172. [Google Scholar]
- Bailey, L. The control of nosema disease. Bee World 1954, 35, 111–113. [Google Scholar]
- Bailey, L. The epidemiology and control of nosema disease of the honey bee. Ann. Appl. Biol. 1955, 43, 379–389. [Google Scholar] [CrossRef]
- Langridge, D.F. Nosema disease of the honeybee and some investigations into its control in Victoria, Australia. Bee World 1961, 42, 36–40. [Google Scholar]
- Fingler, B.D.; Nash, W.T.; Szabo, T.I. A comparison of two techniques for the measurement of Nosema disease in honey bee colonies wintered in Alberta, Canade. Am. Bee J. 1982, 122, 369–371. [Google Scholar]
- Smart, M.D.; Sheppard, W.S. Nosema ceranae in age cohorts of the western honey bee (Apis mellifera). J. Invertebr. Pathol. 2012, 109, 148–151. [Google Scholar] [CrossRef]
- El-Shemy, A.A.M.; Pickard, R.S. Nosema apis Zander infection levels in honeybees of known age. J. Apic. Res. 1989, 28, 101–106. [Google Scholar]
- Morse, R.A.; Flottum, K. Honey Bee Pests, Predators, & Diseases, 3rd ed.; A.I. Root Company: Medina, Saudi Arabia, 1997; p. 718. [Google Scholar]
- Cantwell, G.E. Standard methods for counting Nosema spores. Am. Bee J. 1970, 110, 222–223. [Google Scholar]
- Traver, B.E.; Fell, R.D. Prevalence and infection intensity of Nosema in honey bee (Apis mellifera L.) colonies in Virginia. J. Invertebr. Pathol. 2011, 107, 43–49. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mulholland, G.E.; Traver, B.E.; Johnson, N.G.; Fell, R.D. Individual Variability of Nosema ceranae Infections in Apis mellifera Colonies. Insects 2012, 3, 1143-1155. https://doi.org/10.3390/insects3041143
Mulholland GE, Traver BE, Johnson NG, Fell RD. Individual Variability of Nosema ceranae Infections in Apis mellifera Colonies. Insects. 2012; 3(4):1143-1155. https://doi.org/10.3390/insects3041143
Chicago/Turabian StyleMulholland, Grace E., Brenna E. Traver, Nels G. Johnson, and Richard D. Fell. 2012. "Individual Variability of Nosema ceranae Infections in Apis mellifera Colonies" Insects 3, no. 4: 1143-1155. https://doi.org/10.3390/insects3041143




