Genetic Diversity and Population Structure of Busseola segeta Bowden (Lepidoptera; Noctuidae): A Case Study of Host Use Diversification in Guineo-Congolian Rainforest Relic Area, Kenya
Abstract
:1. Introduction
2. Experimental
2.1. Description of the Study Area
2.2. Sampling, Rearing and Identification of Stem Borers
2.3. DNA Extraction and Sequence Analysis
2.4. Data Management and Statistical Analysis
3. Results
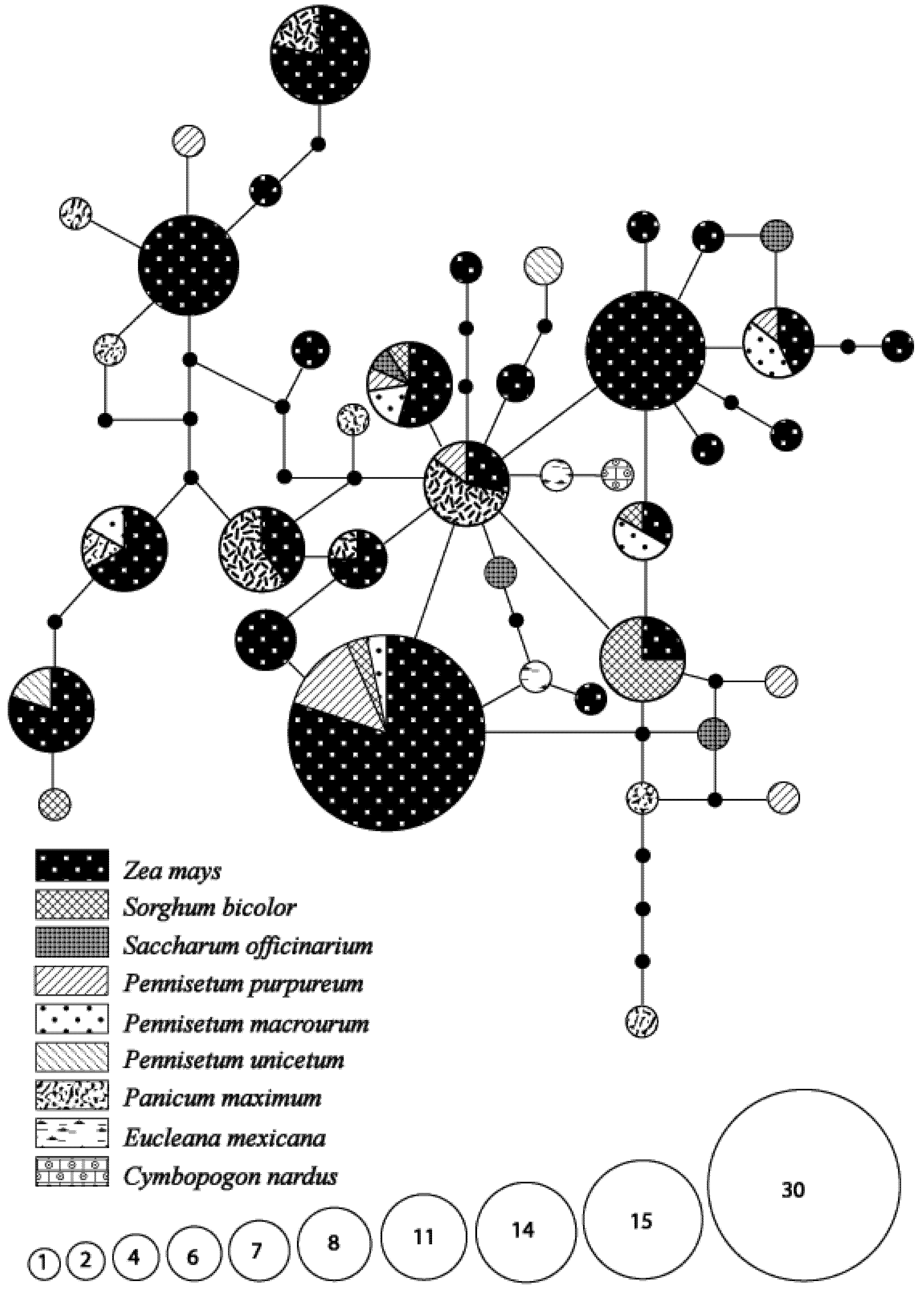
3.1. Species Identity and Host Use Diversity
| Stem borer species | B. fusca (KII) | B. fusca (KI) | S. calamistis (Crop) | S. calamistis (Wild) | M. melanondota | M. nubifera | B. segeta |
|---|---|---|---|---|---|---|---|
| B. fusca KII) | – | – | – | – | – | – | – |
| B. fusca (KI) | 0.026 | – | – | – | – | – | – |
| S. calamistis (Crop) | 0.131 | 0.127 | – | – | – | – | – |
| S. calamistis (Wild) | 0.122 | 0.118 | 0.019 | – | – | – | – |
| M. melanondota | 0.133 | 0.124 | 0.133 | 0.138 | – | – | – |
| M. nubifera | 0.134 | 0.122 | 0.143 | 0.136 | 0.066 | – | – |
| B. segeta | 0.077 | 0.084 | 0.135 | 0.133 | 0.124 | 0.133 | – |
| Host plant species | Total number of larvae recovered | |
|---|---|---|
| Long rain season | Short rain season | |
| Cymbopogon nardus (L.) Rendle | 1 | - |
| Euclaena mexicana Schrader | 2 | - |
| Panicum maximum Jacquin | 28 | 29 |
| Pennisetum macrourum Trinius | 9 | 1 |
| Pennisetum purpureum Schumach* | 18 | 25 |
| Pennisetum unisetum (Nees) Benth. | - | 12 |
| Saccharum officinarum L.* | 2 | 2 |
| Sorghum bicolor Delile* | 18 | - |
| Zea mays L* | 65 | 115 |
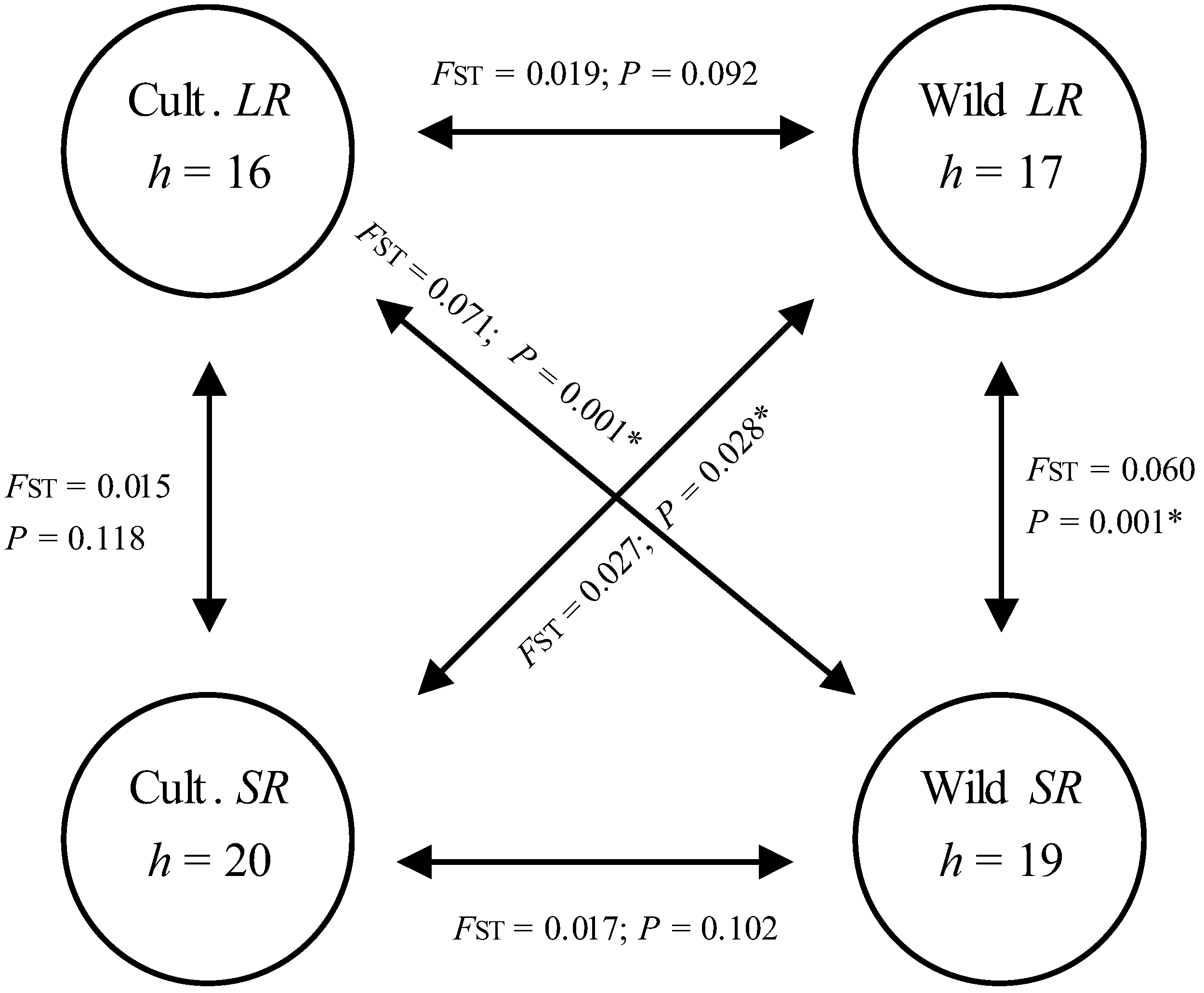
3.2. Genetic Diversity and Differentiation in Host Utilization
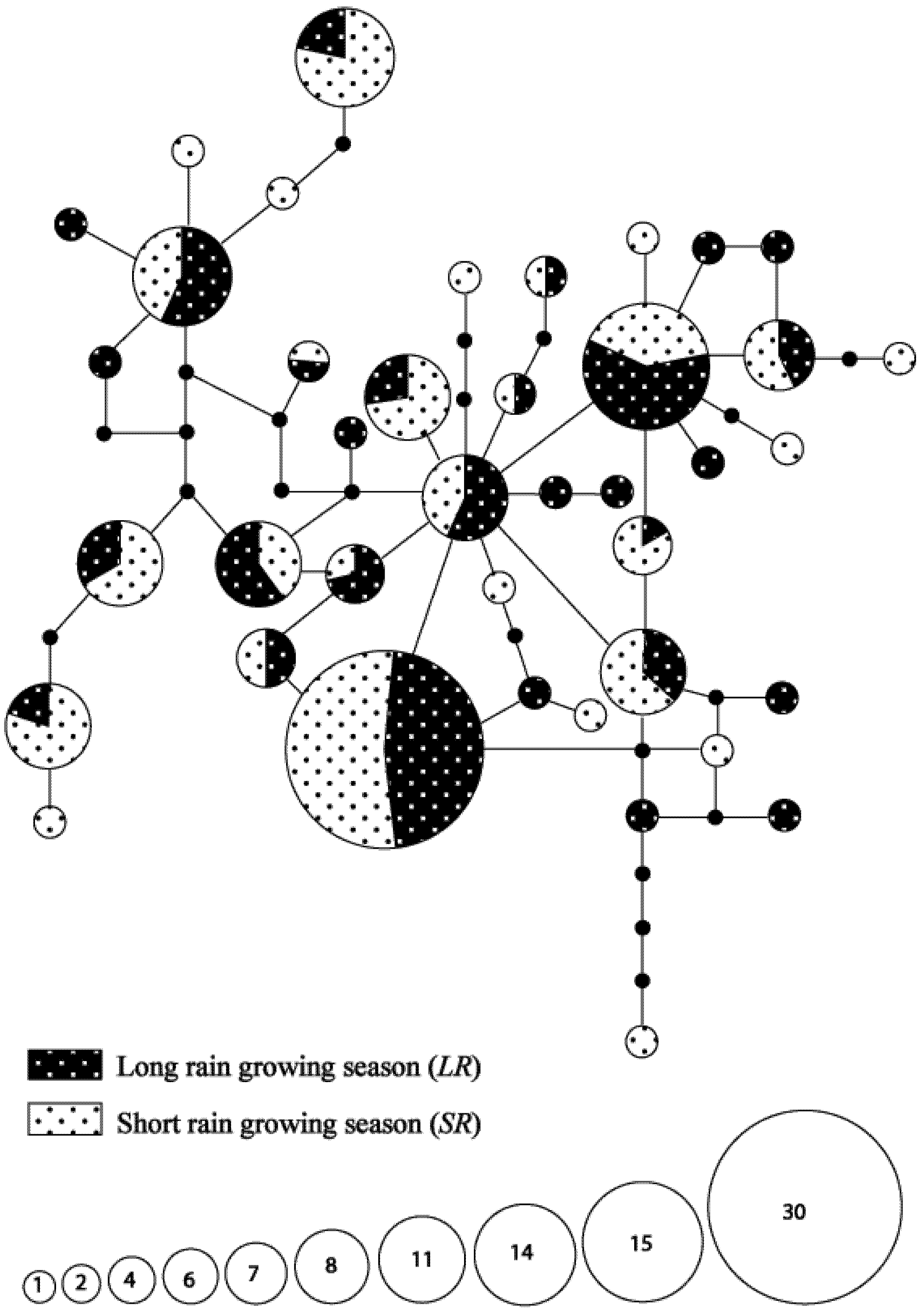
3.3. Seasonal Variations in Haplotype Composition
3.4. Hypothetical Exchange of Haplotypes between Habitats and Seasons
| Genetic parameters | Cultivated host plants | Wild host plants | |||||
|---|---|---|---|---|---|---|---|
| LR | SR | Total | LR | SR | Total | ||
| Number of sequences | 42 | 36 | 78 | 35 | 34 | 69 | |
| Number of segregating sites, S | 20 | 28 | 35 | 25 | 20 | 28 | |
| Number of haplotypes, h | 16 | 20 | 28 | 17 | 19 | 23 | |
| Haplotype diversity, Hd | 0.904 | 0.948 | 0.932 | 0.946 | 0.950 | 0.951 | |
| Average number of differences, K | 3.113 | 3.632 | 3.375 | 3.832 | 3.403 | 3.641 | |
| Nucleotide diversity, Pi | 0.004 | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | |
| AMOVA results | FST | 0.015 | 0.060 | ||||
| p | 0.118 | 0.001 | |||||
4. Discussion
5. Conclusion
Acknowledgements
References
- Bosque-Pérez, N.A.; Schulthess, F. Maize: West and Central Africa. In African Cereal Stem borers: Economic Importance, Taxonomy, Natural Enemies and Control; Polaszek, A., Ed.; CAB International in association with the ACP-EU Technical Centre for Agricultural and Rural Co-operation (CTA): Wageningen, the Netherlands, 1998; pp. 11–24. [Google Scholar]
- Bourguet, D.; Bethenod, M.T.; Trouvé, C.; Viard, F. Host-plant diversity of the European corn borer Ostrinia nubilalis: What value for sustainable transgenic insecticidal Bt corn? P. R. Soc. London B 2000, 267, 1177–1184. [Google Scholar] [CrossRef]
- Gepts, P. Who owns biodiversity and how should the owners be compensated? Plant Physiol. 2004, 134, 1295–1307. [Google Scholar] [CrossRef]
- Ndemah, R.; Schulthess, F.; Le Ru, B.; Bame, I. Lepidopteran cereal stem borers and associated natural enemies on maize and wild grass hosts in Cameroon. J. Appl. Entomol. 2007, 131, 658–668. [Google Scholar] [CrossRef]
- Futuyma, D.J.; Moreno, G. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 1988, 19, 207–233. [Google Scholar]
- Manel, S.; Schwartz, M.K.; Luikart, G.; Taberlet, P. Landscape genetics: Combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Thies, C.; Steffan-Dewenter, I.; Tscharntke, T. Effects of landscape context on herbivory and parasitism at different spatial scales. Oikos 2003, 101, 18–25. [Google Scholar] [CrossRef]
- Gassmann, A.J.; Levy, A.; Tran, T.; Futuyma, D.J. Adaptations of an insect to a novel host plant: A phylogenetic approach. Funct. Ecol. 2006, 20, 478–485. [Google Scholar]
- Moczek, P.A. Developmental capacitance, genetic accommodation, and adaptive evolution. Evol. Dev. 2007, 9, 299–305. [Google Scholar] [CrossRef]
- Polaszek, A.; Khan, Z.R. Host plants. In African Cereal Stem Borers: Economic Importance, Taxonomy, Natural Enemies and Control; Polaszek, A., Ed.; CAB International: Wallingford, UK, 1998; pp. 4–10. [Google Scholar]
- Khan, Z.R.; Midega, C.A.O.; Njuguna, E.M.; Amudavi, D.M.; Wanyama, J.M.; Pickett, J.A. Economic performance of the 'push-pull' technology for stem borer and Striga control in smallholder farming systems in western Kenya. Crop Prot. 2008, 27, 1084–1097. [Google Scholar] [CrossRef]
- Nye, I.W.B. The insect pests of graminaceous crops in East Africa. In Colonial Research Studies No. 31; Her Majesty’s Stationery Office: London, UK, 1960; p. 48. [Google Scholar]
- Le Ru, B.P.; Ong’amo, G.O.; Moyal, P.; Muchugu, E.; Ngala, L.; Musyoka, B.; Abdullah, Z.; Matama-Kauma, T.; Lada, V.Y.; Pallangyo, B.; et al. Geographic distribution and host plant ranges of East African noctuid stem borers. Annales de la Société Entomologique de France (Nouvelle Série) 2006a, 42, 353–361. [Google Scholar]
- Ong’amo, G.O.; Le Ru, B.P.; Dupas, S.; Moyal, P.; Muchugu, E.; Calatayud, P.-A.; Silvain, J.-F. The role of wild host plants in the abundance of lepidopteran stem borers along altitudinal gradient in Kenya. Annales de la Société Entomologique de France (Nouvelle Série) 2006, 42, 363–370. [Google Scholar]
- Ong’amo, G.O. Diversity, Ecology and Population dynamic of Lepidopteran stem borers in Kenya. PhD Thesis, Department of Zoological Sciences, Kenyatta University, Nairobi, Kenya, 2009. [Google Scholar]
- Forman, R.T.T. Land Mosaics:The Ecology of Landscapes and Regions; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Loxdale, H.D.; Lushai, G. Slaves of the environment: the movement of insects in relation to their ecology and genotype. Philos. Trans. R. Soc. B 1999, 354, 1479–1495. [Google Scholar] [CrossRef]
- Gaete-Eastman, C.; Figueroa, C.C.; Olivares-Donoso, R.; Niemeyer, H.M. Diet breadth and its relationship with genetic diversity and differentiation: the case of southern beech aphids (Hemiptera: Aphididae). B. Entomol. Res. 2004, 94, 219–227. [Google Scholar]
- Angelone, S.; Hilfiker, K.; Holderegger, R.; Bergamini, A.; Hoebee, S.E. Regional population dynamics define the local genetic structure in Sorbus torminalis. Mol. Ecol. 2007, 16, 1291–1301. [Google Scholar] [CrossRef]
- Sezonlin, M.; Dupas, S.; Le Ru, B.; Le Gall, P.; Moyal, P.; Calatayud, P.-A.; Giffard, I.; Faure, N.; Silvain, J.-F. Phylogeography and population genetics of the maize stalk borer Busseola fusca (Lepidoptera, Noctuidae) in sub-Saharan Africa. Mol. Ecol. 2006, 15, 407–420. [Google Scholar]
- Lushai, G.; Loxdale, H.D. Tracking movement in small insect pests, with special reference to aphid populations. Int. J. Pest Manage. 2004, 50, 307–315. [Google Scholar] [CrossRef]
- Cords, M.; Tsingalia, H.M. A Report on Harmful Exploitation of Indigenous Forest in Kakamega (to the Ministry of Tourism and Wildlife); Wildlife Conservation and Management Division: Government of Kenya, Nairobi, Kenya, 1982. [Google Scholar]
- Kokwaro, J.O. Conservation status of the Kakamega Forest in Kenya: The easternmost relic of the equatorial rain forests of Africa. Monogr. Syst. Bot. 1988, 25, 471–489. [Google Scholar]
- Tsingalia, M.H. Animals and the Regeneration of an African Rainforest Tree. Ph.D. Thesis, University of California, Berkeley, CA, USA, 1988. [Google Scholar]
- Le Ru, B.P.; Ong’amo, G.O.; Moyal, P.; Ngala, L.; Musyoka, B.; Abdullah, Z.; Cugala, D.; Defabachew, B.; Hailei, T.A.; Kauma, T.M.; et al. Diversity of lepidopteran stem borers in eastern Africa revisited. B. Entomol. Res. 2006B, 96, 555–563. [Google Scholar]
- Gounou, S.; Schulthess, F. Spatial distribution of lepidopterous stem borers on indigenous host plants in West Africa and its implications for sampling schemes. Afr. Entomol. 2004, 12, 171–178. [Google Scholar]
- Onyango, F.O.; Ochieng’ Odero, J.P.R. Continuous rearing of the maize stem borer Busseola fusca on an artificial diet. Entomol. Exp. Appl. 1994, 73, 139–144. [Google Scholar] [CrossRef]
- Harry, M.; Solignac, M.; Lachaise, D. Molecular evidence for parallel evolution of adaptive syndromes in fig-breeding Lissocephala (Drosophilidae). Mol. Phylogenet. Evol. 1998, 9, 542–551. [Google Scholar] [CrossRef]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar]
- Maddison, D.R.; Maddison, W.P. MacClade; Sinauer: Sunderland, MA, USA, 2001. [Google Scholar]
- Rozas, J.; Sànchez-DelBarrio, J.C.; Messeguer, X.; Roza, R. DnaSP DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Lynch, M.; Crease, T.J. The analysis of population survey data on DNA sequence variation. Mol. Biol. Evol. 1990, 7, 377–394. [Google Scholar]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In H.N. Munro, Mammalian Protein Metabolism; Academic Press: New York, NY, USA, 1969. [Google Scholar]
- Tajima, F. Evolutionary relationship of DNA sequences in finite populations. Genetics 1983, 105, 437–460. [Google Scholar]
- Hudson, R.R.; Slatkin, M.; Maddison, W.P. Estimation of levels of gene flow from DNA sequence data. Genetics 1992, 132, 583–589. [Google Scholar]
- Schneider, S.D.; Roessli, D.; Excoffier, L. ARLEQUIN (version 2,000): A Software for Genetic Data Analysis; Genetics and Biometry Laboratory, University of Geneva: Geneva, Switzerland, 2000. [Google Scholar]
- Otieno, N.A.; Le Rü, B.P.; Ong’amo, G.O.; Moyal, P.; Dupas, S.; Calatayud, P.-A.; Silvain, J.-F. Diversity and abundance of wild host plants of lepidopteran stem borers in two agro-ecological zones of Kenya. Int. J. Biodivers. Sci. Manage. 2008, 4, 1–12. [Google Scholar] [CrossRef]
- Tischendorf, L.; Bender, D.J.; Fahrig, L. Evaluation of patch isolation metrics in mosaic landscapes for specialist vs. generalist dispersers. Landscape Ecol. 2003, 18, 41–50. [Google Scholar] [CrossRef]
- Vialatte, A.; Dedryver, C.A.; Simon, J.C.; Galman, M.; Plantegenest, M. Limited genetic exchanges between populations of an insect pest living on uncultivated and related cultivated host plants. P. R. Soc. London B 2005, 272, 1075–1082. [Google Scholar] [CrossRef]
- Mazodze, R.; Conlong, D.E. Eldana saccharina (Lepidoptera: Pyralidae) in sugarcane (Saccharum hybrids), sedges (Cyperus digitatus) and bulrush (Typha latifolia) in south-eastern Zimbabwe. P. S. Afr. Sug. 2003, 77, 256–274. [Google Scholar]
- Futuyma, D.J. Macroevolutionary consequences of speciation: Inferences from phytophagous insects. In Speciation and Its Consequences; Otte, D., Endler, J.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1989; pp. 557–578. [Google Scholar]
- Futuyma, D.J. Evolutionary Biology; Sinauer Associates: Sunderland, MA, USA, 1986. [Google Scholar]
- Ehrlich, P.R.; Raven, P.H. Butterflies and plants: A study in coevolution. Evolution 1964, 18, 586–608. [Google Scholar] [CrossRef]
- Mitter, C.; Farrel, B.; Wiegmann, B. The phylogenetic study of adaptive zones: Has phytophagy promoted insect diversification? Am. Nat. 1988, 132, 107–128. [Google Scholar]
- Ong’amo, G.O.; Le Ru, B.P.; Moyal, P.; Calatayud, P.-A.; Le Gall, P.; Ogol, C.K.P.O.; Kokwaro, E.D.; Capdevielle-Dulac, C.; Silvain, J.-F. Host-plant diversity of Sesamia calamistis: cytochrome b gene sequences reveal local genetic differentiation. Entomol. Exp. Appl. 2008, 128, 154–161. [Google Scholar] [CrossRef]
- Nylin, S.; Gotthard, K. Plasticity in life-history traits. Annu. Rev. Entomol. 1998, 43, 63–83. [Google Scholar] [CrossRef]
- Hawthorne, D.J.; Via, S. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 2001, 412, 904–907. [Google Scholar] [CrossRef]
- Janz, N.; Nyblom, K.; Nylin, S. Evolutionary dynamics of host-plant specialization: A case study of the tribe Nymphalini. Evolution 2001, 55, 783–796. [Google Scholar] [CrossRef]
- Kennedy, G.G.; Storer, N.P. Life system of polyphagous arthropod pests in temporarily unstable cropping systems. Annu. Rev. Entomol. 2000, 45, 467–493. [Google Scholar] [CrossRef]
- West-Eberhard, M.-J. Developmental Plasticity and Evolution; Oxford University Press: New York, NY, USA, 2003. [Google Scholar]
- Overholt, W.A.; Ochieng, J.O.; Lammers, P.M.; Ogedah, K. Rearing and field release methods for Cotesia flavipes Cameron (Hymenoptera: Braconidae), a parasitoid of tropical gramineous stemborers. Insect Sci. Appl. 1994, 15, 253–259. [Google Scholar]
- Schulthess, F.; Bosque-Perez, N.A.; Chabi-Olaye, A.; Gounou, S.; Ndemah, R.; Goergen, G. Exchange of Natural enemies of lepidopteran cereal stem borers between African regions. Insect Sci. Appl. 1997, 17, 97–108. [Google Scholar]
- Ndemah, R.; Gounou, S.; Schulthess, F. The role of wild grasses in the management of lepidopterous cereal stemborers in the humid tropics of western Africa. B. Entomol. Res. 2002, 92, 507–519. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ong'amo, G.O.; Ru, B.P.L.; Campagne, P.; Branca, A.; Calatayud, P.-A.; Capdevielle-Dulac, C.; Silvain, J.-F. Genetic Diversity and Population Structure of Busseola segeta Bowden (Lepidoptera; Noctuidae): A Case Study of Host Use Diversification in Guineo-Congolian Rainforest Relic Area, Kenya. Insects 2012, 3, 1156-1170. https://doi.org/10.3390/insects3041156
Ong'amo GO, Ru BPL, Campagne P, Branca A, Calatayud P-A, Capdevielle-Dulac C, Silvain J-F. Genetic Diversity and Population Structure of Busseola segeta Bowden (Lepidoptera; Noctuidae): A Case Study of Host Use Diversification in Guineo-Congolian Rainforest Relic Area, Kenya. Insects. 2012; 3(4):1156-1170. https://doi.org/10.3390/insects3041156
Chicago/Turabian StyleOng'amo, George O., Bruno P. Le Ru, Pascal Campagne, Antoine Branca, Paul-Andre Calatayud, Claire Capdevielle-Dulac, and Jean-Francois Silvain. 2012. "Genetic Diversity and Population Structure of Busseola segeta Bowden (Lepidoptera; Noctuidae): A Case Study of Host Use Diversification in Guineo-Congolian Rainforest Relic Area, Kenya" Insects 3, no. 4: 1156-1170. https://doi.org/10.3390/insects3041156




