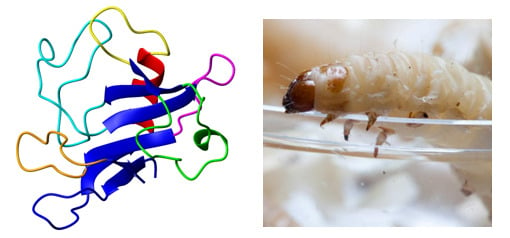
Hirsutellin A: A Paradigmatic Example of the Insecticidal Function of Fungal Ribotoxins
Abstract
:1. Introduction
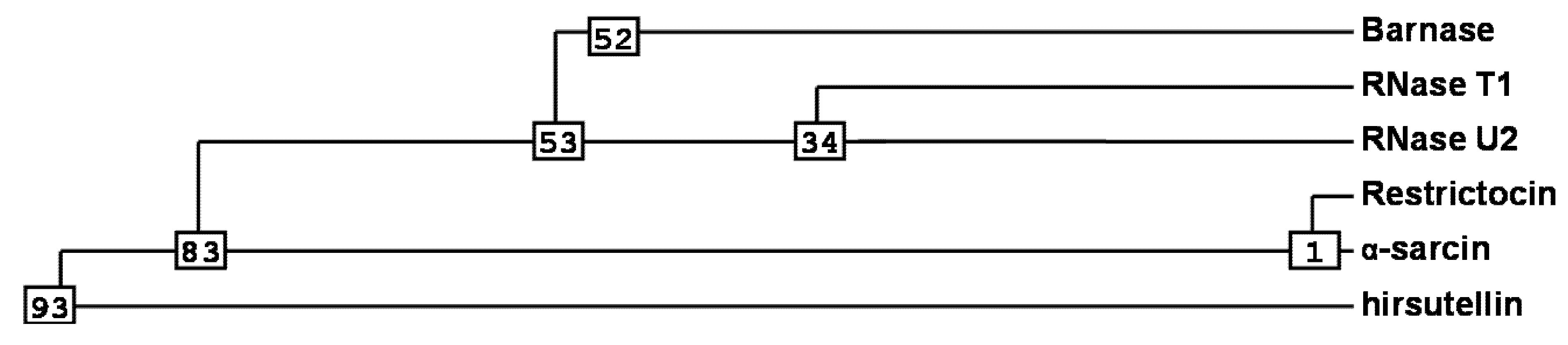
2. Ribotoxins Evolution: Role of HtA
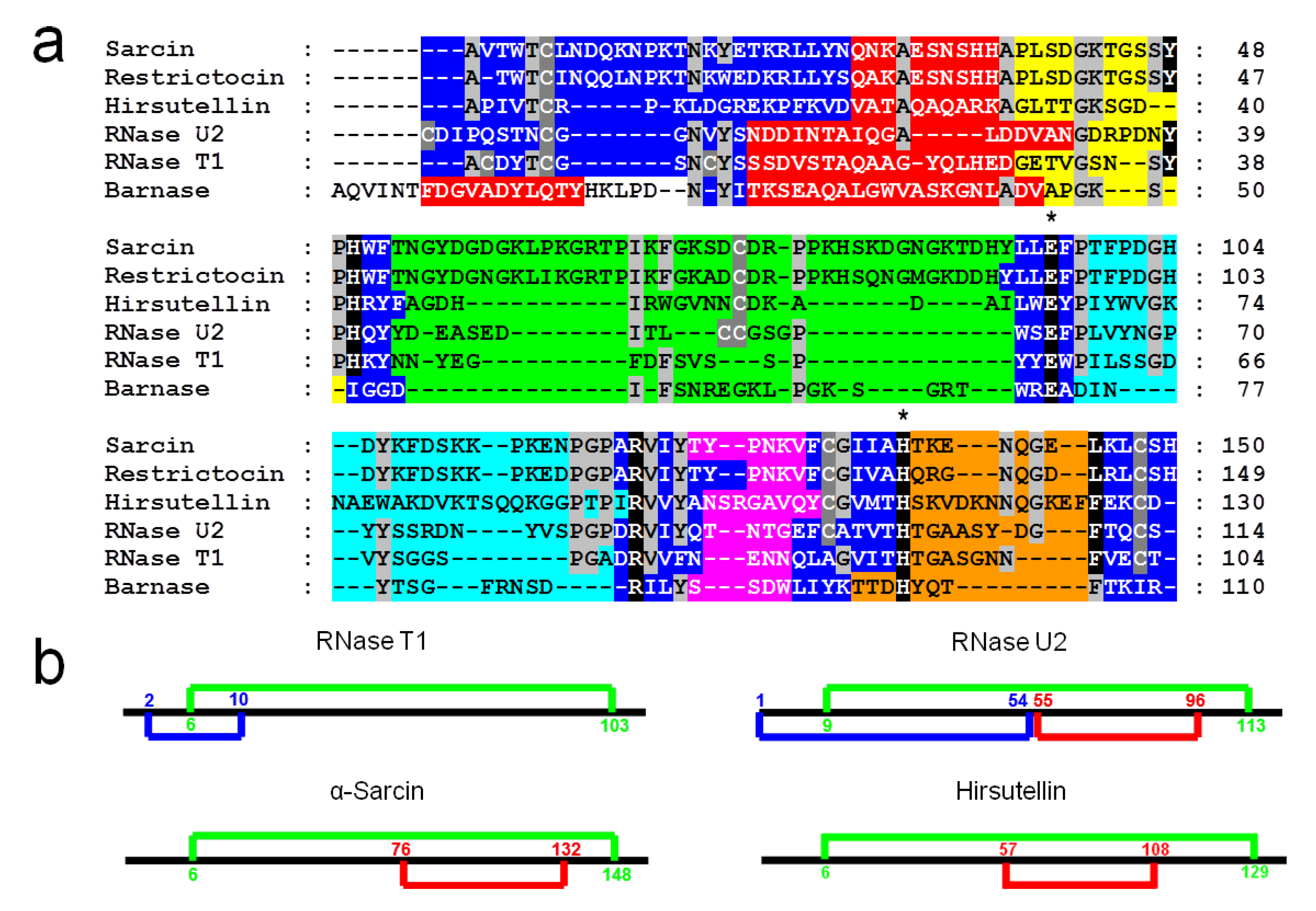
3. Structural Features
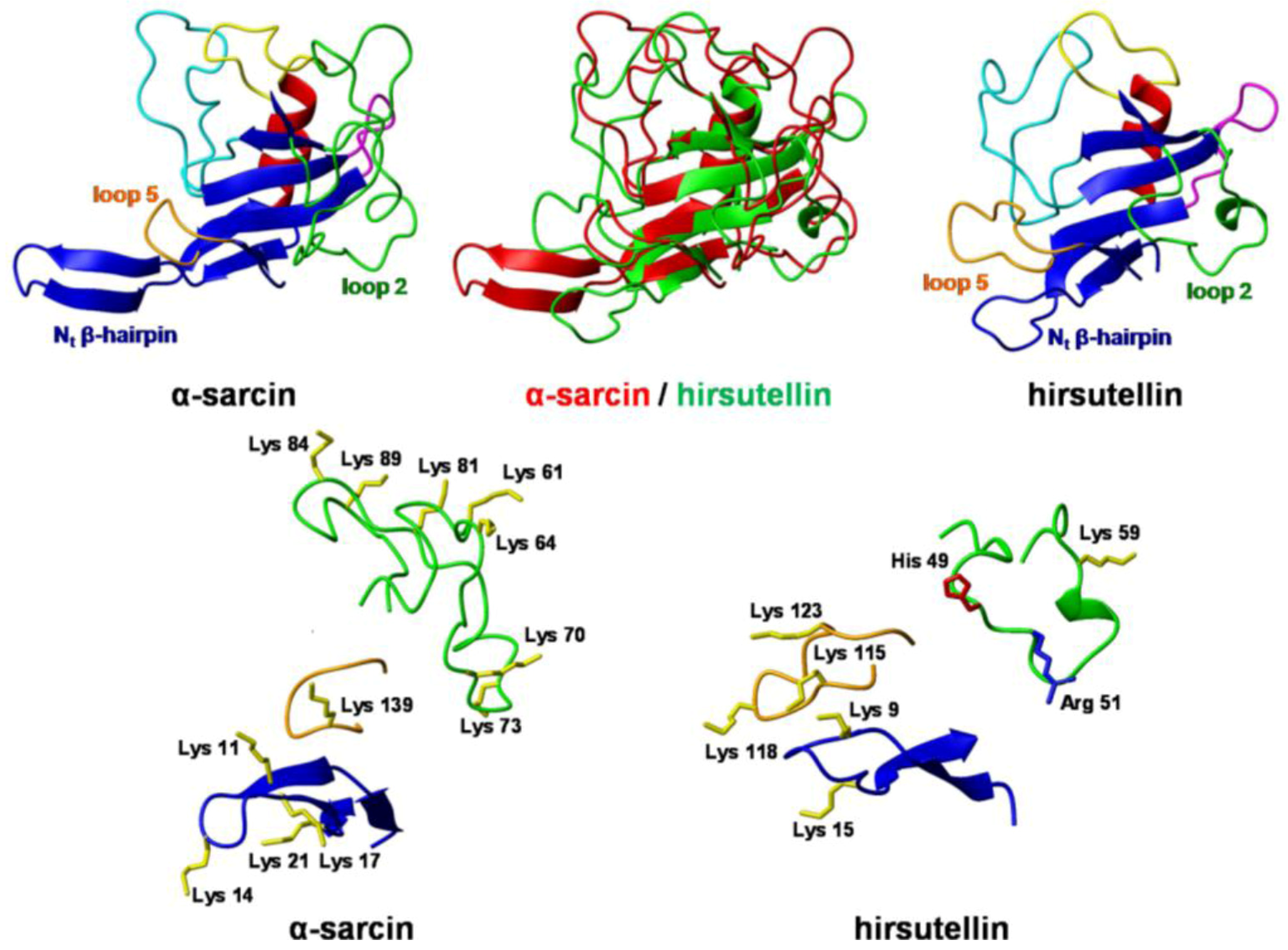

4. Functional Properties
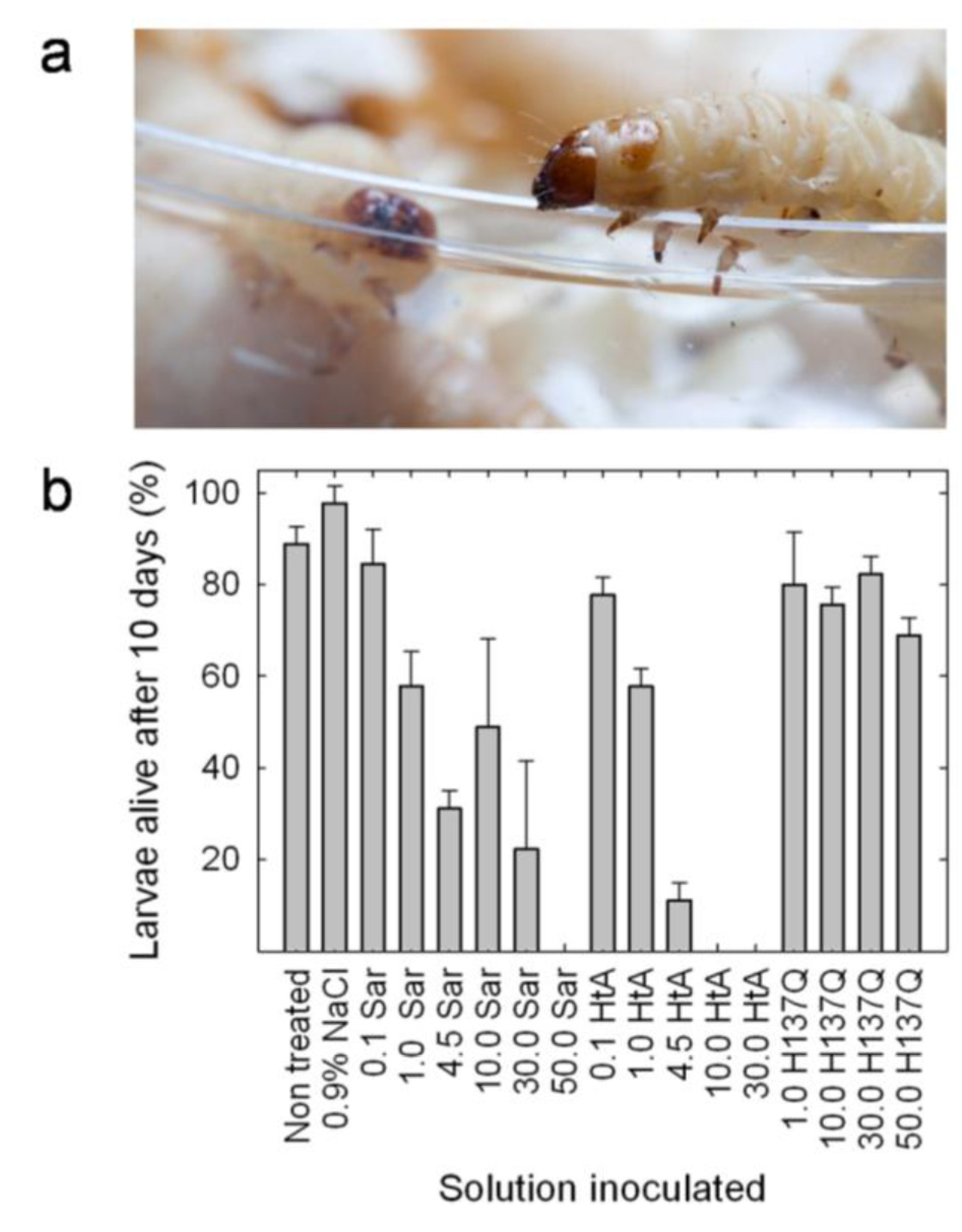
5. Insecticidal Activity

| Viability | Protein biosynthesis inhibition | ||
|---|---|---|---|
| Sf9 | High Five | Sf9 | |
| 0.010 µM | 0.100 µM | 0.04 µM | |
| HtA | 0.010 µM | 0.025 µM | 0.07 µM |
6. Conclusion
Supplementary Files
Supplementary File 1Acknowledgments
Conflict of Interest
References and Notes
- Berenbaum, M.R.; Eisner, T. Bugs' bugs. Science 2008, 322, 52–53. [Google Scholar] [CrossRef]
- Boddy, L.; Jones, T.H. Interactions between basidiomycota and invertebrates. In Ecology of saprotrophic basidiomycetes, Proceedings of the British Mycological Society Symposia, Manchester, UK, September 7th 2007; Boddy, L., Frankland, J.C., van West, P., Eds.; Academic Press: Amsterdam, Nederland, 2008; pp. 155–179. [Google Scholar]
- Ruess, L.; Lussenhop, J. Trophic interactions of fungi and animals. In The fungal community—its organization and role in the ecosystem; Dighton, J., White, F.J., Oudemans, P., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 581–598. [Google Scholar]
- Hegedus, N.; Marx, F. Antifungal proteins: More than antimicrobials? Fungal Biol. Rev. 2013, 26, 132–145. [Google Scholar] [CrossRef]
- Olson, B.H.; Goerner, G.L. Alpha-sarcin, a new antitumor agent I. Isolation, purification, chemical composition, and the identity of a new amino acid. Appl. Microbiol. 1965, 13, 314–321. [Google Scholar]
- Endo, Y.; Wool, I.G. The site of action of α-sarcin on eukaryotic ribosomes. The sequence at the α-sarcin cleavage site in 28S ribosomal ribonucleic acid. J. Biol. Chem. 1982, 257, 9054–9060. [Google Scholar]
- Schindler, D.G.; Davies, J.E. Specific cleavage of ribosomal RNA caused by α-sarcin. Nucleic Acids Res. 1977, 4, 1097–1110. [Google Scholar] [CrossRef]
- Gasset, M.; Mancheño, J.M.; Lacadena, J.; Turnay, J.; Olmo, N.; Lizarbe, M.A.; Martinez-del-Pozo, A.; Oñaderra, M.; Gavilanes, J.G. α-Sarcin, a ribosome-inactivating protein that translocates across the membranes of phospholipid vesicles. Curr. Topics. Pept. Membr. Res. 1994, 1, 99–104. [Google Scholar]
- Lacadena, J.; Alvarez-Garcia, E.; Carreras-Sangra, N.; Herrero-Galan, E.; Alegre-Cebollada, J.; Garcia-Ortega, L.; Oñaderra, M.; Gavilanes, J.G.; Martinez-del-Pozo, A. Fungal ribotoxins: Molecular dissection of a family of natural killers. FEMS Microbiol. Rev. 2007, 31, 212–237. [Google Scholar] [CrossRef]
- Martinez-Ruiz, A.; Kao, R.; Davies, J.; Martinez-del-Pozo, A. Ribotoxins are a more widespread group of proteins within the filamentous fungi than previously believed. Toxicon 1999, 37, 1549–1563. [Google Scholar] [CrossRef]
- Chan, Y.L.; Endo, Y.; Wool, I.G. The sequence of the nucleotides at the α-sarcin cleavage site in rat 28S ribosomal ribonucleic acid. J. Biol. Chem. 1983, 258, 12768–12770. [Google Scholar]
- Endo, Y.; Huber, P.W.; Wool, I.G. The ribonuclease activity of the cytotoxin α-sarcin. The characteristics of the enzymatic activity of alpha-sarcin with ribosomes and ribonucleic acids as substrates. J. Biol. Chem. 1983, 258, 2662–2667. [Google Scholar]
- Olmo, N.; Turnay, J.; Gonzalez de Buitrago, G.; Lopez de Silanes, I.; Gavilanes, J.G.; Lizarbe, M.A. Cytotoxic mechanism of the ribotoxin α-sarcin. Induction of cell death via apoptosis. Eur. J. Biochem. 2001, 268, 2113–2123. [Google Scholar] [CrossRef]
- Alvarez-Garcia, E.; Batanero, E.; Garcia-Fernandez, R.; Villalba, M.; Gavilanes, J.G.; Martinez-del-Pozo, A. A deletion variant of the Aspergillus fumigatus ribotoxin Aspf1 induces an attenuated airway inflammatory response in a mouse model of sensitization. J. Investig. Allergol. Clin. Immunol. 2010, 20, 69–75. [Google Scholar]
- Arruda, L.K.; Mann, B.J.; Chapman, M.D. Selective expression of a major allergen and cytotoxin, Aspf1, in Aspergillus fumigatus. Implications for the immunopathogenesis of Aspergillus-related diseases. J. Immunol. 1992, 149, 3354–3359. [Google Scholar]
- Roga, V.; Hedeman, L.P.; Olson, B.H. Evaluation of mitogillin (NSC-69529) in the treatment of naturally occurring canine neoplasms. Cancer Chemother. Rep. 1971, 55, 101–113. [Google Scholar]
- Brandhorst, T.; Dowd, P.F.; Kenealy, W.R. The ribosome-inactivating protein restrictocin deters insect feeding on Aspergillus restrictus. Microbiology 1996, 142, 1551–1556. [Google Scholar] [CrossRef]
- Brandhorst, T.; Dowd, P.F.; Kenealy, W.R. The effect of fungal ribosome inactivating proteins upon feeding choice in C. freemani, and indications of a mutualistic relationship with A. restrictus. Mycopathologia 2001, 152, 155–158. [Google Scholar] [CrossRef]
- Brandhorst, T.T.; Kenealy, W.R. Production and localization of restrictocin in Aspergillus restrictus. J. Gen. Microbiol. 1992, 138, 1429–1435. [Google Scholar] [CrossRef]
- Yang, R.; Kenealy, W.R. Regulation of restrictocin production in Aspergillus restrictus. J. Gen. Microbiol. 1992, 138, 1421–1427. [Google Scholar] [CrossRef]
- Lamy, B.; Davies, J. Isolation and nucleotide sequence of the Aspergillus restrictus gene coding for the ribonucleolytic toxin restrictocin and its expression in Aspergillus nidulans: The leader sequence protects producing strains from suicide. Nucleic Acids Res. 1991, 19, 1001–1006. [Google Scholar] [CrossRef]
- Yang, R.; Kenealy, W.R. Effects of amino-terminal extensions and specific mutations on the activity of restrictocin. J. Biol. Chem. 1992, 267, 16801–16805. [Google Scholar]
- Krasnoff, S.B.; Gupta, S. Identification of the antibiotic phomalactone from the entomopathogenic fungus, Hirsutella thompsonii var. synnematosa. J. Chem. Ecol. 1994, 293–302. [Google Scholar] [CrossRef]
- Vey, A.; Quiot, J.M.; Mazet, I.; McCoy, C.W. Toxicity and pathology of crude broth filtarte produced by Hirsutella thompsonii var. thompsonii in shake culture. J. Invertebr. Pathol. 1993, 61, 131–137. [Google Scholar] [CrossRef]
- Samson, R.A.; McCoy, C.W.; O'Donnell, K.L. Taxonomy of the acarine parasite Hirsutella thompsonii. Mycologia 1980, 72, 359–377. [Google Scholar] [CrossRef]
- Omoto, C.; McCoy, C.W. Toxicity of purified fungal toxin hirsutellin A to the citrus rust mite Phyllocoptruta oleivora (ash.). J. Invertebr. Pathol. 1998, 72, 319–322. [Google Scholar] [CrossRef]
- Liu, J.C.; Boucias, D.G.; Pendland, J.C.; Liu, W.-Z.; Maruniak, J. The mode of action of hirsutellin A on eukaryotic cells. J. Invertebr. Pathol. 1996, 67, 224–228. [Google Scholar] [CrossRef]
- Liu, W.Z.; Boucias, D.G.; McCoy, C.W. Extraction and characterization of the insecticidal toxin hirsutellin A produced by Hirsutella thompsonii var. thompsonii. Exp. Mycol. 1995, 19, 254–262. [Google Scholar] [CrossRef]
- Mazet, I.; Vey, A. Hirsutellin A, a toxic protein produced in vitro by Hirsutella thompsonii. Microbiology 1995, 141, 1343–1348. [Google Scholar] [CrossRef]
- Herrero-Galan, E.; Lacadena, J.; Martinez-del-Pozo, A.; Boucias, D.G.; Olmo, N.; Oñaderra, M.; Gavilanes, J.G. The insecticidal protein hirsutellin A from the mite fungal pathogen Hirsutella thompsonii is a ribotoxin. Proteins 2008, 72, 217–228. [Google Scholar]
- Olombrada, M.; Herrero-Galan, E.; Tello, D.; Oñaderra, M.; Gavilanes, J.G.; Martinez-del-Pozo, A.; Garcia-Ortega, L. Fungal extracellular ribotoxins as insecticidal agents. Insect Biochem. Mol. Biol. 2013, 43, 39–46. [Google Scholar] [CrossRef]
- Kao, R.; Davies, J. Fungal ribotoxins: A family of naturally engineered targeted toxins? Biochem. Cell. Biol. 1995, 73, 1151–1159. [Google Scholar] [CrossRef]
- Lamy, B.; Davies, J.; Schindler, D. The Aspergillus ribonucleolytic toxins (ribotoxins). In Genetically Engineered Toxins; Frankel, A.E., Ed.; Marcel Dekker Inc.: New York, NY, USA, 1992; pp. 237–258. [Google Scholar]
- Aravind, L.; Koonin, E.V. A natural classification of ribonucleases. Methods Enzymol. 2001, 341, 3–28. [Google Scholar] [CrossRef]
- Perez-Cañadillas, J.M.; Santoro, J.; Campos-Olivas, R.; Lacadena, J.; Martinez-del-Pozo, A.; Gavilanes, J.G.; Rico, M.; Bruix, M. The highly refined solution structure of the cytotoxic ribonuclease α-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity. J. Mol. Biol. 2000, 299, 1061–1073. [Google Scholar] [CrossRef]
- Martinez-Ruiz, A.; Garcia-Ortega, L.; Kao, R.; Lacadena, J.; Oñaderra, M.; Mancheño, J.M.; Davies, J.; Martinez-del-Pozo, A.; Gavilanes, J.G. RNase U2 and α-sarcin: A study of relationships. Methods Enzymol. 2001, 341, 335–351. [Google Scholar] [CrossRef]
- Martinez-Ruiz, A.; Martinez-del-Pozo, A.; Lacadena, J.; Oñaderra, M.; Gavilanes, J.G. Hirsutellin A displays significant homology to microbial extracellular ribonucleases. J. Invertebr. Pathol. 1999, 74, 96–97. [Google Scholar] [CrossRef]
- Martinez-del-Pozo, A.; Gasset, M.; Oñaderra, M.; Gavilanes, J.G. Conformational study of the antitumor protein α-sarcin. Biochim. Biophys. Acta 1988, 953, 280–288. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Barber, D.; Fernandez-Luna, J.L.; Soriano, F.; Mendez, E. The primary structure of the cytotoxin restrictocin. Eur. J. Biochem. 1984, 143, 621–634. [Google Scholar] [CrossRef]
- Sacco, G.; Drickamer, K.; Wool, I.G. The primary structure of the cytotoxin α-sarcin. J. Biol. Chem. 1983, 258, 5811–5818. [Google Scholar]
- Viegas, A.; Herrero-Galan, E.; Oñaderra, M.; Macedo, A.L.; Bruix, M. Solution structure of hirsutellin A: new insights into the active site and interacting interfaces of ribotoxins. FEBS J. 2009, 276, 2381–2390. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Yang, X.; Gerczei, T.; Glover, L.; Correll, C.C. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat. Struct. Biol. 2001, 8, 968–973. [Google Scholar] [CrossRef]
- Campos-Olivas, R.; Bruix, M.; Santoro, J.; Martinez-del-Pozo, A.; Lacadena, J.; Gavilanes, J.G.; Rico, M. H-1 and N-15 nuclear magnetic resonance assignment and secondary structure of the cytotoxic ribonuclease α-sarcin. Protein Sci. 1996, 5, 969–972. [Google Scholar]
- Garcia-Mayoral, M.F.; Garcia-Ortega, L.; Lillo, M.P.; Santoro, J.; Martinez-del-Pozo, A.; Gavilanes, J.G.; Rico, M.; Bruix, M. NMR structure of the noncytotoxic α-sarcin mutant ∆(7-22): The importance of the native conformation of peripheral loops for activity. Protein Sci. 2004, 13, 1000–1011. [Google Scholar] [CrossRef]
- Garcia-Mayoral, M.F.; Perez-Cañadillas, J.M.; Santoro, J.; Ibarra-Molero, B.; Sanchez-Ruiz, J.M.; Lacadena, J.; Martinez-del-Pozo, A.; Gavilanes, J.G.; Rico, M.; Bruix, M. Dissecting structural and electrostatic interactions of charged groups in α-sarcin. An NMR study of some mutants involving the catalytic residues. Biochemistry 2003, 42, 13122–13133. [Google Scholar] [CrossRef]
- Boucias, D.G.; Farmerie, W.G.; Pendland, J.C. Cloning and sequencing of cDNA of the insecticidal toxin hirsutellin A. J. Invertebr. Pathol. 1998, 72, 258–261. [Google Scholar] [CrossRef]
- Garcia-Ortega, L.; Lacadena, J.; Villalba, M.; Rodriguez, R.; Crespo, J.F.; Rodriguez, J.; Pascual, C.; Olmo, N.; Oñaderra, M.; Martinez-del-Pozo, A.; et al. Production and characterization of a noncytotoxic deletion variant of the Aspergillus fumigatus allergen Aspf1 displaying reduced IgE binding. FEBS J. 2005, 272, 2536–2544. [Google Scholar] [CrossRef]
- Koradi, R.; Billeter, M.; Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Herrero-Galan, E.; Garcia-Ortega, L.; Lacadena, J.; Martinez-del-Pozo, A.; Olmo, N.; Gavilanes, J.G.; Oñaderra, M. Implication of an Asp residue in the ribonucleolytic activity of hirsutellin A reveals new electrostatic interactions at the active site of ribotoxins. Biochimie 2012, 94, 427–433. [Google Scholar] [CrossRef]
- Herrero-Galan, E.; Garcia-Ortega, L.; Lacadena, J.; Martinez-del-Pozo, A.; Olmo, N.; Gavilanes, J.G.; Oñaderra, M. A non-cytotoxic but ribonucleolytically specific ribotoxin variant: Implication of tryptophan residues in the cytotoxicity of hirsutellin A. Biol. Chem. 2012, 393, 449–456. [Google Scholar]
- Mancheño, J.M.; Gasset, M.; Albar, J.P.; Lacadena, J.; Martínez del Pozo, A.; Oñaderra, M.; Gavilanes, J.G. Membrane interaction of a beta-structure-forming synthetic peptide comprising the 116–139th sequence region of the cytotoxic protein α-sarcin. Biophys. J. 1995, 68, 2387–2395. [Google Scholar] [CrossRef]
- Lacadena, J.; Martinez-del-Pozo, A.; Lacadena, V.; Martinez-Ruiz, A.; Mancheño, J.M.; Oñaderra, M.; Gavilanes, J.G. The cytotoxin α-sarcin behaves as a cyclizing ribonuclease. FEBS Lett. 1998, 424, 46–48. [Google Scholar] [CrossRef]
- Garcia-Ortega, L.; Alvarez-Garcia, E.; Gavilanes, J.G.; Martinez-del-Pozo, A.; Joseph, S. Cleavage of the sarcin-ricin loop of 23S rRNA differentially affects EF-G and EF-Tu binding. Nucleic Acids Res. 2010, 38, 4108–4119. [Google Scholar] [CrossRef]
- Alvarez-Garcia, E.; Garcia-Ortega, L.; Verdun, Y.; Bruix, M.; Martinez-del-Pozo, A.; Gavilanes, J.G. Tyr-48, a conserved residue in ribotoxins, is involved in the RNA-degrading activity of α-sarcin. Biol. Chem. 2006, 387, 535–541. [Google Scholar]
- Lacadena, J.; Mancheño, J.M.; Martinez-Ruiz, A.; Martinez-del-Pozo, A.; Gasset, M.; Oñaderra, M.; Gavilanes, J.G. Substitution of histidine-137 by glutamine abolishes the catalytic activity of the ribosome-inactivating protein α-sarcin. Biochem. J. 1995, 309, 581–586. [Google Scholar]
- Lacadena, J.; Martinez-del-Pozo, A.; Martinez-Ruiz, A.; Perez-Cañadillas, J.M.; Bruix, M.; Mancheño, J.M.; Oñaderra, M.; Gavilanes, J.G. Role of histidine-50, glutamic acid-96, and histidine-137 in the ribonucleolytic mechanism of the ribotoxin α-sarcin. Proteins 1999, 37, 474–484. [Google Scholar] [CrossRef]
- Masip, M.; Garcia-Ortega, L.; Olmo, N.; Garcia-Mayoral, M.F.; Pérez-Cañadillas, J.M.; Bruix, M.; Oñaderra, M.; Martinez-del-Pozo, A.; Gavilanes, J.G. Leucine 145 of the ribotoxin α-sarcin plays a key role for determining the specificity of the ribosome-inactivating activity of the protein. Protein Sci. 2003, 12, 161–169. [Google Scholar] [CrossRef]
- Masip, M.; Lacadena, J.; Mancheño, J.M.; Oñaderra, M.; Martinez-Ruiz, A.; Martinez-del-Pozo, A.; Gavilanes, J.G. Arginine 121 is a crucial residue for the specific cytotoxic activity of the ribotoxin α-sarcin. Eur. J. Biochem. 2001, 268, 6190–6196. [Google Scholar] [CrossRef]
- Siemer, A.; Masip, M.; Carreras, N.; Garcia-Ortega, L.; Oñaderra, M.; Bruix, M.; Martinez-del-Pozo, A.; Gavilanes, J.G. Conserved asparagine residue 54 of α-sarcin plays a role in protein stability and enzyme activity. Biol. Chem. 2004, 385, 1165–1170. [Google Scholar]
- Pérez-Cañadillas, J.M.; Campos-Olivas, R.; Lacadena, J.; Martinez-del-Pozo, A.; Gavilanes, J.G.; Santoro, J.; Rico, M.; Bruix, M. Characterization of pK(a) values and titration shifts in the cytotoxic ribonuclease α-sarcin by NMR. Relationship between electrostatic interactions, structure, and catalytic function. Biochemistry 1998, 37, 15865–15876. [Google Scholar] [CrossRef]
- Turnay, J.; Olmo, N.; Jimenez, A.; Lizarbe, M.A.; Gavilanes, J.G. Kinetic study of the cytotoxic effect of α-sarcin, a ribosome inactivating protein from Aspergillus giganteus, on tumour cell lines: Protein biosynthesis inhibition and cell binding. Mol. Cell. Biochem. 1993, 122, 39–47. [Google Scholar] [CrossRef]
- Di Donato, A.; Cafaro, V.; Dalessio, G. Ribonuclease A can be transformed into a dimeric ribonuclease with antitumor activity. J. Biol. Chem. 1994, 269, 17394–17396. [Google Scholar]
- Ilinskaya, O.N.; Dreyer, F.; Mitkevich, V.A.; Shaw, K.L.; Pace, C.N.; Makarov, A.A. Changing the net charge from negative to positive makes ribonuclease Sa cytotoxic. Protein Sci. 2002, 11, 2522–2525. [Google Scholar]
- Vatzaki, E.H.; Allen, S.C.; Leonidas, D.D.; Trautwein-Fritz, K.; Stackhouse, J.; Benner, S.A.; Acharya, K.R. Crystal structure of a hybrid between ribonuclease A and bovine seminal ribonuclease—the basic surface, at 2.0 Å resolution. Eur. J. Biochem. 1999, 260, 176–182. [Google Scholar] [CrossRef]
- Carreras-Sangra, N.; Alvarez-Garcia, E.; Herrero-Galan, E.; Tome-Amat, J.; Lacadena, J.; Alegre-Cebollada, J.; Oñaderra, M.; Gavilanes, J.G.; Martinez-del-Pozo, A. The therapeutic potential of fungal ribotoxins. Curr. Pharm. Biotechnol. 2008, 9, 153–160. [Google Scholar] [CrossRef]
- Gasset, M.; Martinez-del-Pozo, A.; Oñaderra, M.; Gavilanes, J.G. Study of the interaction between the antitumour protein α-sarcin and phospholipid vesicles. Biochem. J. 1989, 258, 569–575. [Google Scholar]
- Gasset, M.; Oñaderra, M.; Martinez-del-Pozo, A.; Schiavo, G.-P.; Laynez, J.; Usobiaga, P.; Gavilanes, J.G. Effect of the antitumour protein α-sarcin on the thermotropic behaviour of acid phospholipid vesicles. Biochim. Biophys. Acta 1991, 1068, 9–16. [Google Scholar]
- Gasset, M.; Oñaderra, M.; Goormaghtigh, E.; Gavilanes, J.G. Acid phospholipid vesicles produce conformational changes on the antitumour protein α-sarcin. Biochim. Biophys. Acta 1991, 1080, 51–58. [Google Scholar]
- Gasset, M.; Oñaderra, M.; Thomas, P.G.; Gavilanes, J.G. Fusion of phospholipid vesicles produced by the anti-tumour protein α-sarcin. Biochem.J. 1990, 265, 815–822. [Google Scholar]
- Mancheño, J.M.; Gasset, M.; Lacadena, J.; Ramon, F.; Martinez-del-Pozo, A.; Oñaderra, M.; Gavilanes, J.G. Kinetic study of the aggregation and lipid mixing produced by α-sarcin on phosphatidylglycerol and phosphatidylserine vesicles: Stopped-flow light scattering and fluorescence energy transfer measurements. Biophys. J. 1994, 67, 1117–1125. [Google Scholar] [CrossRef]
- Bergelson, L.D.; Dyatlovitskaya, E.V.; Torkhovskaya, T.I.; Sorokina, I.B.; Gorkova, N.P. Phospholipid composition of membranes in the tumor cell. Biochim. Biophys. Acta 1970, 210, 287–298. [Google Scholar] [CrossRef]
- Carrasco, L.; Esteban, M. Modification of membrane permeability in vaccinia virus-infected cells. Virology 1982, 117, 62–69. [Google Scholar] [CrossRef]
- Fernandez-Puentes, C.; Carrasco, L. Viral infection permeabilizes mammalian cells to protein toxins. Cell 1980, 20, 769–775. [Google Scholar]
- Orntoft, T.F.; Vestergaard, E.M. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis 1999, 20, 362–371. [Google Scholar] [CrossRef]
- Ran, S.; Downes, A.; Thorpe, P.E. Increased exposure of anionic phospholipids on the surface of tumor blood vessels. Cancer Res. 2002, 62, 6132–6140. [Google Scholar]
- Connor, J.; Bucana, C.; Fidler, I.J.; Schroit, A.J. Differentiation-dependent expression of phosphatidylserine in mammalian plasma membranes: Quantitative assessment of outer-leaflet lipid by prothrombinase complex formation. Proc. Natl. Acad. Sci. USA 1989, 86, 3184–3188. [Google Scholar] [CrossRef]
- Zachowsky, A. Phospholipids in animal eukaryotic membranes: Transverse asymmetry and movement. Biochem. J. 1993, 294, 1–14. [Google Scholar]
- Filigheddu, N.; Cutrupi, S.; Porporato, P.E.; Riboni, F.; Baldanzi, G.; Chianale, F.; Fortina, E.; Piantanida, P.; De Bortoli, M.; Vacca, G.; et al. Diacylglycerol kinase is required for HGF-induced invasiveness and anchorage-independent growth of MDA-MB-231 breast cancer cells. Anticancer Res. 2007, 27, 1489–1492. [Google Scholar]
- Griner, E.M.; Kazanietz, M.G. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer 2007, 7, 281–294. [Google Scholar] [CrossRef]
- Merida, I.; Avila-Flores, A.; Merino, E. Diacylglycerol kinases: At the hub of cell signalling. Biochem. J. 2008, 409, 1–18. [Google Scholar] [CrossRef]
- Garcia-Ortega, L.; Lacadena, J.; Mancheño, J.M.; Oñaderra, M.; Kao, R.; Davies, J.; Olmo, N.; Martinez-del-Pozo, A.; Gavilanes, J.G. Involvement of the N-terminal β-hairpin of the Aspergillus ribotoxins on the interaction with membranes and nonspecific ribonuclease activity. Protein Sci. 2001, 10, 1658–1668. [Google Scholar] [CrossRef]
- Garcia-Ortega, L.; Masip, M.; Mancheño, J.M.; Oñaderra, M.; Lizarbe, M.A.; Garcia-Mayoral, M.F.; Bruix, M.; Martinez-del-Pozo, A.; Gavilanes, J.G. Deletion of the N-terminal β-hairpin of the ribotoxin α-sarcin produces a nontoxic but active ribonuclease. J. Biol. Chem. 2002, 277, 18632–18639. [Google Scholar] [CrossRef]
- Carreras-Sangra, N.; Tome-Amat, J.; Garcia-Ortega, L.; Batt, C.A.; Oñaderra, M.; Martinez-del-Pozo, A.; Gavilanes, J.G.; Lacadena, J. Production and characterization of a colon cancer-specific immunotoxin based on the fungal ribotoxin α-sarcin. Protein Eng. Des. Sel. 2012, 25, 425–435. [Google Scholar] [CrossRef]
- Tome-Amat, J.; Menendez-Mendez, A.; Garcia-Ortega, L.; Batt, C.A.; Oñaderra, M.; Martinez-del-Pozo, A.; Gavilanes, J.G.; Lacadena, J. Production and characterization of scFvA33T1, an immunoRNase targeting colon cancer cells. FEBS J. 2012, 279, 3022–3032. [Google Scholar] [CrossRef]
- Kanga, L.H.; James, R.R.; Boucias, D.G. Hirsutella thompsonii and Metarhizium anisopliae as potential microbial control agents of Varroa destructor, a honey bee parasite. J. Invertebr. Pathol. 2002, 81, 175–184. [Google Scholar] [CrossRef]
- Peng, C.Y.; Zhou, X.; Kaya, H.K. Virulence and site of infection of the fungus Hirsutella thompsonii to the honey bee ectoparasitic mite, Varroa destructor. J. Invertebr. Pathol. 2002, 81, 185–195. [Google Scholar] [CrossRef]
- Marheineke, K.; Grunewald, S.; Christie, W.; Reilander, H. Lipid composition of Spodoptera frugiperda (Sf9) and Trichoplusia ni (Tn) insect cells used for baculovirus infection. FEBS Lett. 1998, 441, 49–52. [Google Scholar] [CrossRef]
- Soderhall, K.; Cerenius, L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr. Opin. Immunol. 1998, 10, 23–28. [Google Scholar] [CrossRef]
- Craik, D.J. Host-defense activities of cyclotides. Toxins 2012, 4, 139–156. [Google Scholar] [CrossRef]
- Windley, M.J.; Herzig, V.; Dziemborowicz, S.A.; Hardy, M.C.; King, G.F.; Nicholson, G.M. Spider-venom peptides as bioinsecticides. Toxins 2012, 4, 191–227. [Google Scholar]
- Rathore, D.; Batra, J.K. Cytotoxic activity of ribonucleolytic toxin restrictocin-based chimeric toxins targeted to epidermal growth factor receptor. FEBS Lett. 1997, 407, 275–279. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Herrero-Galán, E.; García-Ortega, L.; Olombrada, M.; Lacadena, J.; Del Pozo, Á.M.; Gavilanes, J.G.; Oñaderra, M. Hirsutellin A: A Paradigmatic Example of the Insecticidal Function of Fungal Ribotoxins. Insects 2013, 4, 339-356. https://doi.org/10.3390/insects4030339
Herrero-Galán E, García-Ortega L, Olombrada M, Lacadena J, Del Pozo ÁM, Gavilanes JG, Oñaderra M. Hirsutellin A: A Paradigmatic Example of the Insecticidal Function of Fungal Ribotoxins. Insects. 2013; 4(3):339-356. https://doi.org/10.3390/insects4030339
Chicago/Turabian StyleHerrero-Galán, Elías, Lucía García-Ortega, Miriam Olombrada, Javier Lacadena, Álvaro Martínez Del Pozo, José G. Gavilanes, and Mercedes Oñaderra. 2013. "Hirsutellin A: A Paradigmatic Example of the Insecticidal Function of Fungal Ribotoxins" Insects 4, no. 3: 339-356. https://doi.org/10.3390/insects4030339