2. The Cyst, Which Includes a Filamentous Cell, Is the Characteristic and Diagnostic Feature for the Genus Helicosporidium
Virtually all reports of
Helicosporidium infection in invertebrate hosts rely on the observation of unique and characteristic four-cell structures [
1,
3,
4,
5,
6,
7,
8,
9,
10,
11,
12,
13]. These structures have been alternatively termed spores [
1,
3,
4,
5,
6,
7,
9] or cysts [
8,
10,
11,
12,
13]. Although the term spore was used during the original description of
Helicosporidium parasiticum [
1], and can still be found in occasional modern reports [
9], most current studies, including this review, refer to the diagnostic feature of
Helicosporidium as cyst. The
Helicosporidium cyst is a barrel-shape structure that contains a core of three superposed ovoid cells surrounded by a single elongated, filamentous cell. These four cells are enclosed in a pellicle. The original
H. parasiticum description featured elaborate drawings and microphotographs of the cyst and the filamentous cell [
1]. Since then, electron microscopy photographs of the characteristic cyst have routinely been provided to support the identification of
Helicosporidium in insect and other invertebrates. Recent reports of
Helicosporidium sp. in various Coleopteran hosts, including the great European spruce bark beetle
Dendroctonus micans (Coleoptera: Curculionidae), the predator beetle
Rhizophagus grandis (Coleoptera: Rhizophaginae), and the weevil
Cyrtobagous salviniae (Coleoptera: Curculionidae), all incorporated transmission electron micrographs (TEM) depicting cysts with the peripheral filamentous cell surrounding the three ovoid cells [
10,
11,
12]. Cell measurements [
7,
8,
10,
12] indicated that the cysts are rather small, and range from 3 to 6 μm, although some of this variation might be due to differences in preparation methods [
7]. Inside the cysts, the filamentous cell typically wraps around the core of ovoid cells three or four times, and can be distinguished on the narrow outer surface of the cysts (
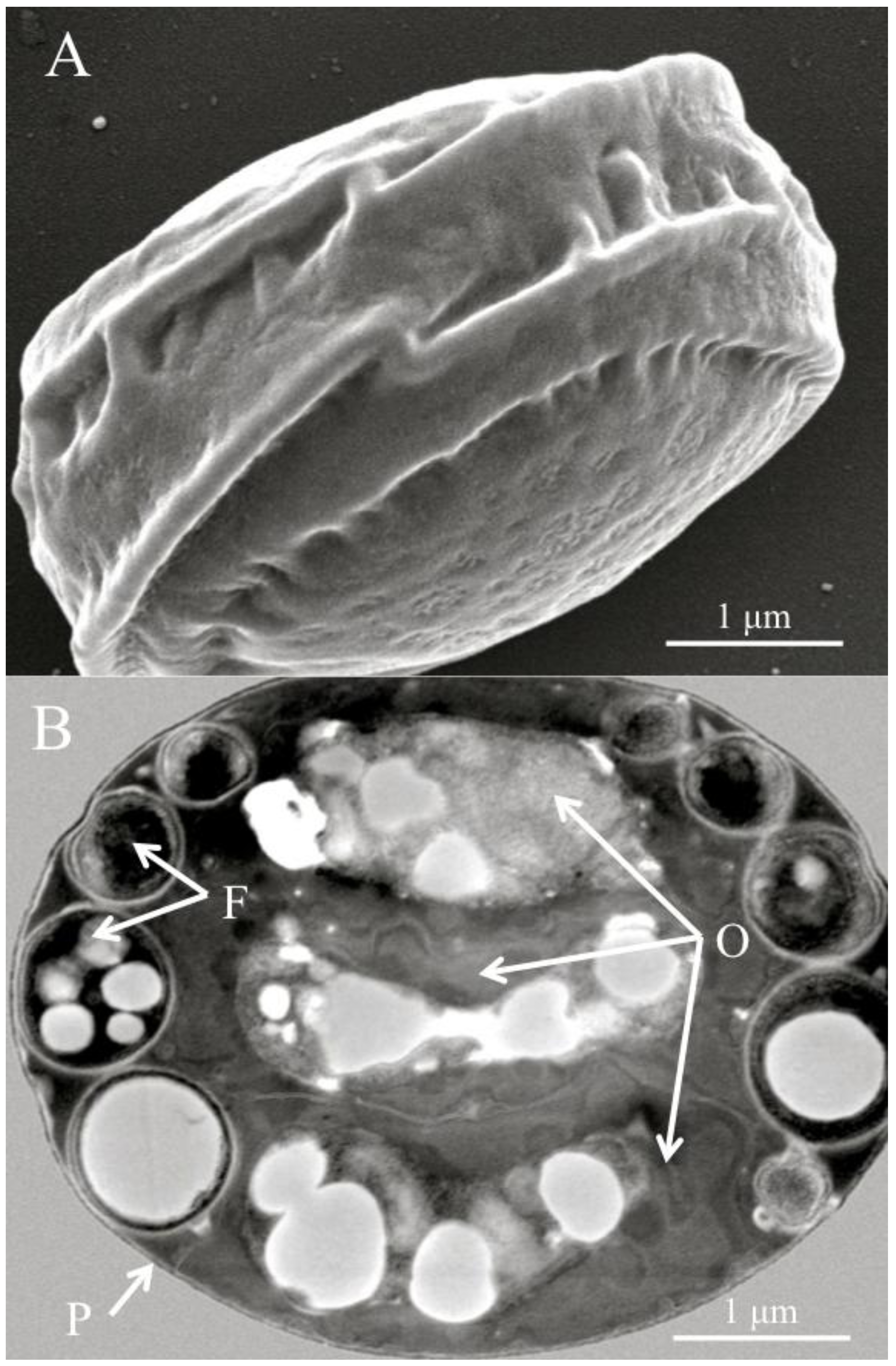
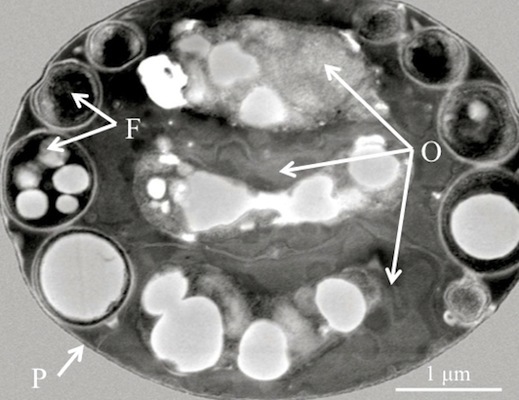
Figure 1).
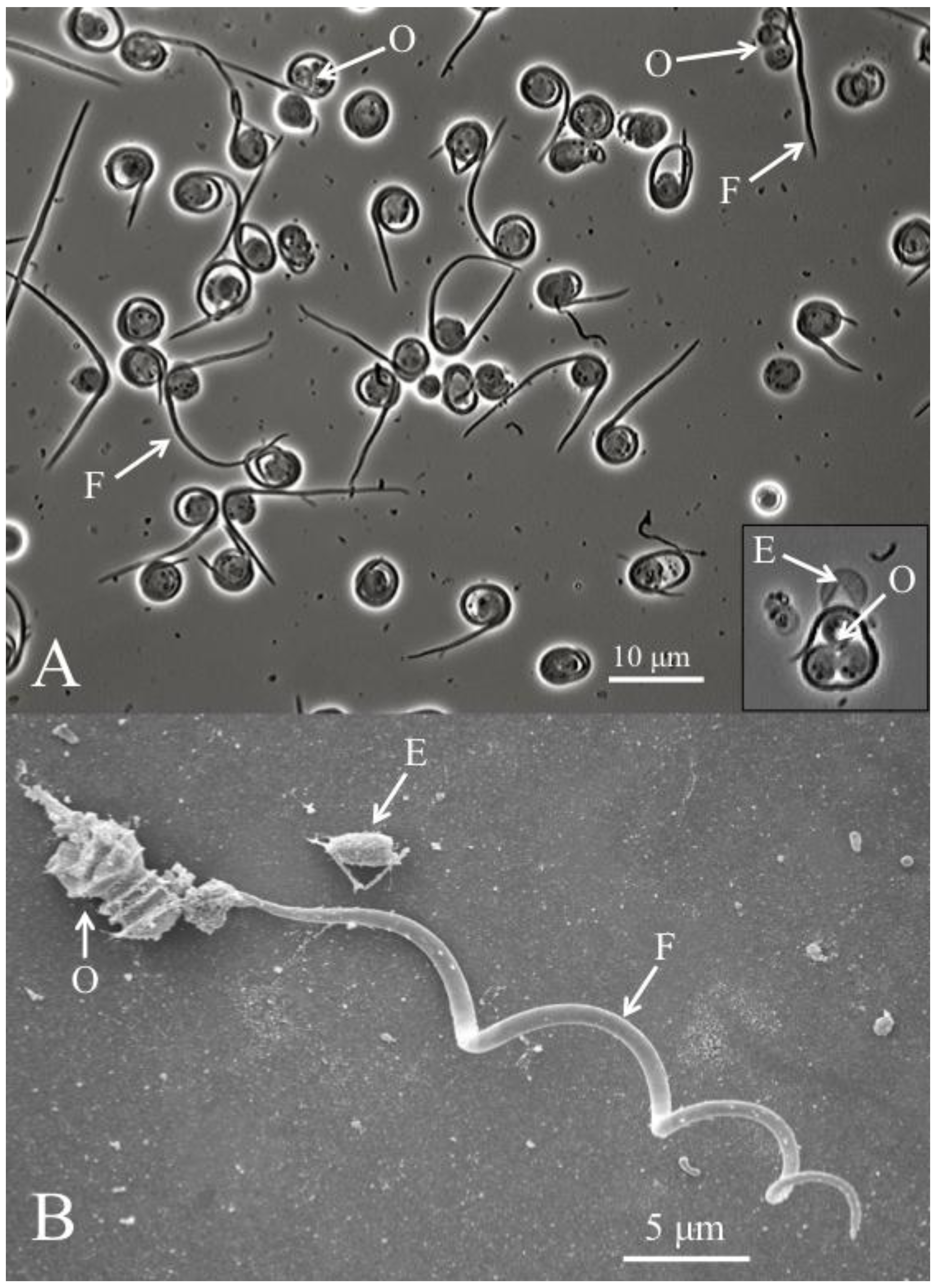
In addition to TEM pictures, a recent study presented scanning electron microscopes pictures of cysts that were purified using Ludox gradient centrifugation, as well as light microscopy and SEM pictures of the filamentous cell being liberated from the cyst and separated from the remaining three ovoid cells [
8]. This process is known as dehiscence (
Figure 2). It has been observed both
in vitro, by applying pressure to the microscope slide [
8], and
in vivo, in the gut lumen of susceptible hosts [
14], and in the host hemolymph, after a series of desiccation events [
1]. Significantly, dehisced cysts were used to single out the filamentous cells and highlight the presence of barbs [
8] (
Figure 2). Purified filamentous cells range in length from 37 to 62 μm [
1,
4,
8,
10,
14]. It remains unclear if these ultrastructural differences can be related to the hosts from which these
Helicosporidium were isolated.
The dividing stage of
Helicosporidium corresponds to vegetative cells. These cells are characterized by the mitotic production of multi-cellular structures containing two, four or eight cells within a pellicle [
14,
15,
16]. Similar to the cyst, the structure can rupture and release daughter cells from an empty pellicle. This mode of reproduction was termed autosporulation in reference to a similar process described for related taxa [
16]. Meiosis has yet to be reported. Vegetative growth have been successfully obtained
in vitro using cell-free artificial media, demonstrating that
Helicosporidium can replicate without living host cells [
8]. In heterologous, lepidopteran hosts, only a minority (15%–21%) of four-cell vegetative cells from a dipteran isolate developed into the typical cysts [
16], and it is unknown if cyst morphogenesis is more efficient in natural hosts. Cyst differentiation has yet to be observed in
in vitro cultures, suggesting that host–derived stimuli are required for this process [
16]. Rather than developing into cysts,
Helicosporidium cells allowed to undergo repetitive autosporulation cycles in artificial media were observed to form palmelloid colonies, which correspond to the agglutination of potentially abnormal vegetative cells [
16]. Another notable difference between
in vitro and
in vivo vegetative development of
Helicosporidium cells involves cell concentrations, as
in vitro cultures obtained in artificial media failed to match the cell density observed in the host hemolymph [
16].
Following the development of axenic cultures [
8], three strains have been deposited the American Type Culture Collection (ATCC). These strains, labeled Sj-1, Dm-1, and Cs-1, refer to isolates, respectively collected from the black fly
Simulium jonesi, the bark beetle
Dendroctonus micans, and the weevil
Cyrtobagous salviniae [
8,
10,
12]. All three strains have been prudently named
Helicosporidium sp., without a full species name, since it remains unclear if the genus is comprised of only one species, and if all
Helicosporidium isolates are
Helicosporidium parasiticum Keilin [
1]. Comparative analyses between isolates are rare and have been complicated by difficulties in locating the original specimen. Hence, the vast majority of new
Helicosporidium reports have been based on the limited description of the diagnostic cysts and filamentous cells, with little attempts to further the identification past the genus level, leading to reports of
Helicosporidium sp. [
6,
7,
8,
9,
10,
11,
12,
13]. Polymorphic traits associated with cell morphology have been identified, but it is unclear if they are taxonomically significant [
8,
10,
17]. Potentially,
in vitro cultures of
Helicosporidium may provide a basis to develop comparative analyses between isolates, and identify differences justifying the description of several
Helicosporidium species. Comparative analyses of
in vitro cultures of
Helicosporidium Sj-1 and related taxa have already been performed [
18]. In addition, axenic cultures will facilitate the characterization of the different cell phenotypes at the molecular level. In particular, the compositions of the
Helicosporidium pellicle, or the vegetative cell wall, remain largely unknown, even if these structures play an important role in pathogenicity. Although molecular data has been generated in the form of expressed sequence tags (ESTs), they have been mainly used for phylogenetic analyses and not for molecular characterization of
Helicosporidium cells [
19]. To date,
in vitro cultures have mostly proved crucial to obtain molecular sequences that precisely established the taxonomic affinities of
Helicosporidium spp., and to investigate the transitions between cell types in order to fully understand the
Helicosporidium life cycle.
Figure 1.
(A) Scanning electron micrograph of a diagnostic Helicosporidium cyst. (B) Transmission electron micrograph (cross section) detailing the core of three stacked ovoid cells (O) and the filamentous cell (F) contained within a pellicle (P).
Figure 1.
(A) Scanning electron micrograph of a diagnostic Helicosporidium cyst. (B) Transmission electron micrograph (cross section) detailing the core of three stacked ovoid cells (O) and the filamentous cell (F) contained within a pellicle (P).
Figure 2.
(A) Helicosporidium cyst dehiscence observed under light microscopy, showing groups of three ovoid cells (O), diagnostic filamentous cells (F) and empty pellicles (E). (B) Scanning electron micrograph detailing the filamentous cell and its barbs (pointing away from the core of ovoid cells).
Figure 2.
(A) Helicosporidium cyst dehiscence observed under light microscopy, showing groups of three ovoid cells (O), diagnostic filamentous cells (F) and empty pellicles (E). (B) Scanning electron micrograph detailing the filamentous cell and its barbs (pointing away from the core of ovoid cells).
3. The Cyst, and the Included Filamentous Cell, Initiates the Helicosporidium Infectious Cycle in Susceptible Insect Hosts
The pathogenicity process of
Helicosporidium spp. has been inferred principally from two long‑term studies that detailed the infection sequence in heterologous lepidopteran hosts maintained in laboratories [
8,
14,
15,
16,
20]. A
Helicosporidium originally isolated from larvae and adults of
Carpophilus mutilatus (Coleoptera, Nitidulidae) was tracked in the navel orangeworm
Paramyelois transitella [
14,
15]. Later,
Helicosporidium Sj-1 was used to obtain infections in common laboratory insects such as the corn earworm
Helicoverpa zea, the tobacco hornworm
Manduca sexta, or the beet armyworm
Spodoptera exigua [
8,
16,
20]. Both studies established that cysts are the infective propagules and that infection occurs
per os, when cysts are ingested by susceptible hosts. During insect bioassays, significant infection rates have routinely be obtained after
per os challenge, even though a significant portion of intact cysts may be retrieved in the animal’s feces [
20]. Gut dissections clearly demonstrated that cyst dehiscence was induced in the gut lumen [
8,
14,
20]. The impact of insect gut on cysts was confirmed
in vitro, as cysts incubated in midgut fluids were induced to rupture and release the filamentous cells [
8]. Ovoid cells contained in the cysts were observed to lyse in the midgut [
8,
20].
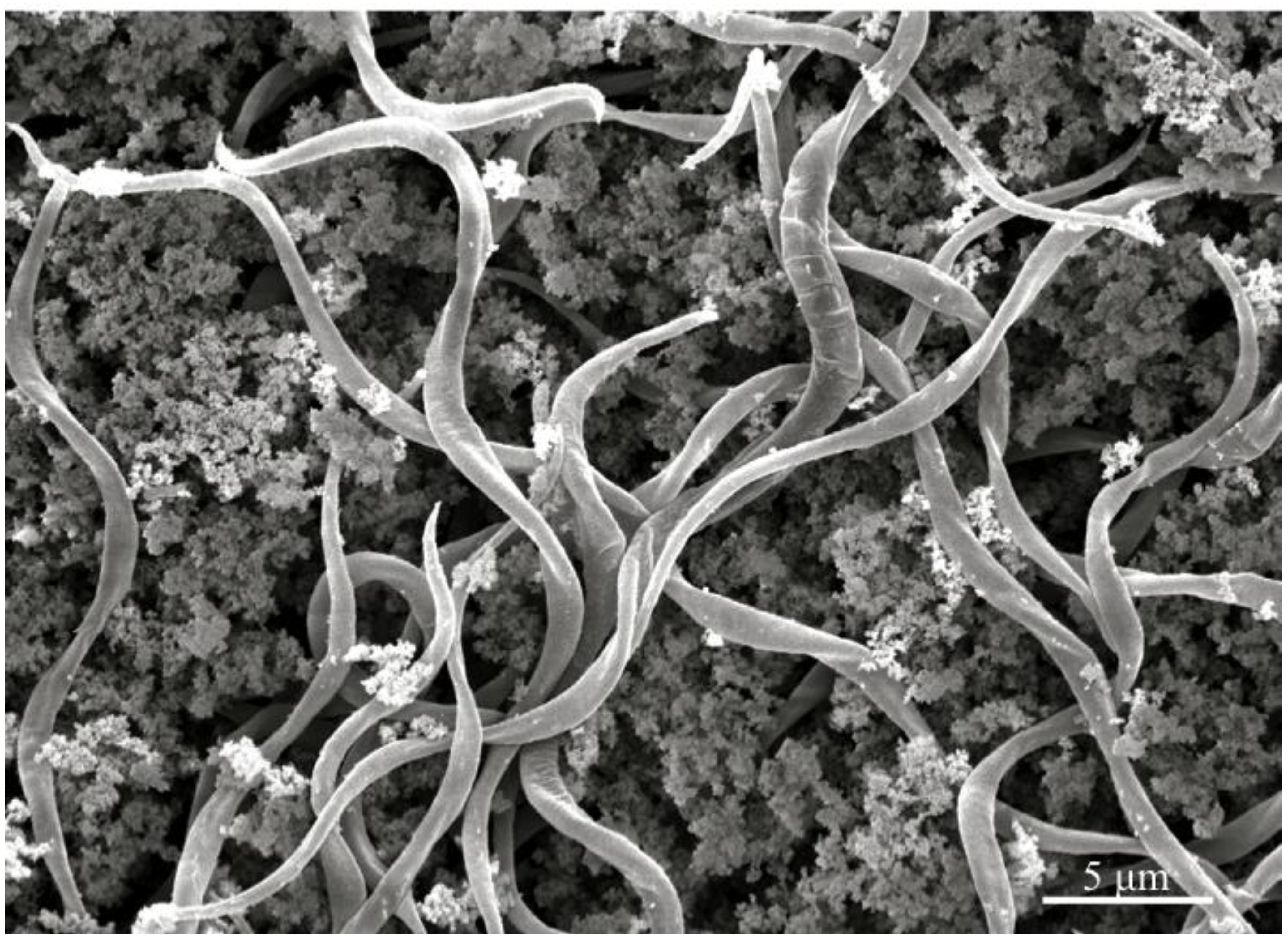
In vivo cyst dehiscence is illustrated in
Figure 3, complementing additional micrographs of insect gut lumen that demonstrated the infection process [
8,
20].
An initial investigation suggested that cysts might bind to the insect peritrophic membrane before dehiscence, and therefore be in a position where the released filamentous cells penetrate the peritrophic matrix immediately after dehiscence [
8]. Although cyst dehiscence was initially described in the host hemocoel [
1], it is now well established that filamentous cells are not found freely circulating in the hemolymph, but mediate the transition from midgut tissues to the host hemolymph, where
Helicosporidium vegetative cells can be observed 24 h or 48 h post infection [
8,
14,
16,
20]. Detailed microscopic observations revealed that the released filamentous cells pass through the midgut epithelium, reach the hemocoel, and may create significant damage to the peritrophic membrane so that remaining cysts and cells gain access to the underlying tissues, and eventually, to the hemocoel [
20]. Even the smallest described filamentous cells (37 μm) were estimated to be long enough to puncture the peritrophic membrane and the ectoperitrophic space, and gain access to the midgut cells [
8]. The orientation of the filamentous cells barbs was found to be highly consistent, suggesting that the host‑pathogen interactions may be highly regulated at the cellular and molecular level. The barbs were first observed to be oriented towards the gut lumen as the filamentous cells initiated contact with the matrix, but then switched orientation and pointed towards the hemocoel as the filamentous cells gain ingress in the hemolymph [
20].
The transition from invasive filamentous cells to replicating vegetative cells was elucidated recently under
in vitro conditions [
16]. In a manner similar to the more commonly observed autosporulation process, filamentous cells swell and undergo two division cycles within the original cell wall, releasing four bean-shaped daughter cells. These elongated cells divide again to produce spherical vegetative cells [
16]. It is assumed that a similar differentiation takes place
in vivo. Although uncommon, elongated cells were observed in both gut and hemolymph preparations of infected
P. transitella [
14]. Remnants of filamentous cells and bean-shaped cells were also observed being phagocytosed in infected
S. exigua hemocytes [
16]. The vegetative cells are responsible for hemolymph colonization. Since no filamentous cells are observed in the hemolymph, it is hypothesized that the transition occurs either in the host phagocytes for the filamentous cells that reach the hemolymph, or during the ingress from the gut tissue to the hemocoel, once the peritrophic matrix has been disrupted [
20].
Figure 3.
Scanning electron micrograph of the gut content of insect hosts challenged with Helicosporidium cysts, demonstrating that filamentous cells are released in the gut lumen.
Figure 3.
Scanning electron micrograph of the gut content of insect hosts challenged with Helicosporidium cysts, demonstrating that filamentous cells are released in the gut lumen.
The vegetative cells are not recognized by the defense system of susceptible hosts, and replicate freely in the hemocoel, or in the hemocytes. They eventually fill the hemocoel, and have been showed to reach very high concentrations [
16,
20]. The
Helicosporidium life cycle culminates as a portion of the vegetative cells differentiate into cysts [
1,
8,
14,
15,
16,
20,
21]. This process has been observed both in the hemolymph and in hemocytes [
20], and is impacted by both host age and pathogen dosage [
21]. Although cyst production has yet to occur in artificial media, the vegetative cells obtained
in vitro were shown to maintain their competence to differentiate into cysts when injected into host hemolymph, suggesting that a host-derived stimulus is required for cyst morphogenesis [
16]. Direct injection of
Helicosporidium cells in host hemolymph also resulted in 100% infection rates, which contrasted with the 50% rates reported for
per os challenges [
22]. This observation not only confirmed that cells are undetected by the host defense system, but also suggested that mechanisms of resistance against
Helicosporidium infection are related to the ingress of the pathogen through the midgut [
22].
High cell concentration and cyst morphogenesis in the hemolymph has variable impact on the insect host. There is no consistent or characteristic symptom associated with
Helicosporidium infection in insects. Noticeable infection symptoms, such as change in hemolymph color and decreased host weight gain [
23], or decreased host mobility [
24], have occasionally been reported. Host death may be considered as the ultimate symptom, as suggested by the original
Helicosporidium description [
1]. However, several studies also indicated that the morphology [
11,
13] and the development (pupation) of infected insects [
9,
17,
22,
25] appeared unaffected. The lack of characteristic symptoms for
Helicosporidium infection is consistent with the variable infectivity and mortality rates reported for this pathogen. Infection rates are dependent on both hosts and strains. A
Helicosporidium sp. isolate tested on various mosquito species resulted in infection rates ranging from 0 to 93% [
17]. In heterologous lepidopteran hosts,
Helicosporidium Cs-1 produced an infection rate of 50% [
22]. Bioassays demonstrated that infection rates are dose dependent [
21,
23]. Mortality rates have been equally variable, and can also be modulated by the host age and the concentration of cysts used to challenge potential hosts [
21,
26]. Significant mortality rates have been reported in several mosquito hosts, spurring substantial investigations on the potential of
Helicosporidium as a biological control agent against mosquito larvae [
9,
17,
24,
25,
26,
27]. Overall, the variability in pathogenicity and virulence observed in different studies may also be correlated to the subsampled diversity within the genus
Helicosporidium. The small number of infection bioassays that attempted to compare several
Helicosporidium isolates all highlighted the fact that distinct isolates can be readily distinguished, based on differences in infectivity or virulence towards a single host [
23,
26]. These observations support the existence of several
Helicosporidium species.
Lastly, although the
Helicosporidium life cycle, from cyst ingestion to cyst morphogenesis in the hemolymph has been clearly established, no study has unambiguously described how
Helicosporidium cysts exit the hemolymph of infected insects and are transmitted to subsequent hosts. The original
H. parasiticum description indicated that infected larvae might become so impaired that the exoskeleton ruptures and the entire hemocoel content (including the infective cysts) is released into the environment [
1]. This release has also been reported in mosquito larvae [
26]. In pathosystems where the hosts appeared mostly unaffected and infection was trans-stadially maintained, vertical transmission of
Helicosporidium was investigated. In mosquito hosts, infection of ovarian tissues was not observed [
25], but vertical transmission was reported in noctuids [
22]. Studies that described infections in both insects of a prey/predator system (the bark beetle
Dendroctonus micans and its predator
Rhizophagus grandis) suggested that
Helicosporidium cysts may be directly transmitted by feeding when insects consume an infected prey, or potentially cannibalize an infected conspecies [
10,
11,
13].
4. Phylogenetic and Phylogenomic Analyses Revealed that Helicosporidium spp. Are Non‑Photosynthetic Green Algae
The taxonomic classification for the genus
Helicosporidium was historically a matter of debate, with these organisms alternatively proposed as Protozoa [
1,
2] or lower Fungi [
3], based on morphological characteristics. However, the phylogenetic affinity of
Helicosporidium has been unequivocally established by molecular-based analyses. The first sequenced gene fragments from
Helicosporidium Sj-1 corresponded to rDNA, actin and β-tubulin. Phylogenetic reconstructions inferred from these genes revealed that members of the genus
Helicosporidium are green algae (Chlorophyta) [
28]. The SSU-rDNA gene fragment was also used to demonstrate that the genus
Helicosporidium belongs to the chlorophyte class Trebouxiophyceae, and is closely related to the non‑photosynthetic algae of the genus
Prototheca [
28]. A set of 69 ribosomal protein sequences from the Sj-1 strain provided strong support, not only for the reclassification of
Helicosporidium as green algae, but also for the specific relationship with
Prototheca spp. [
19]. Following this strongly supported, molecular-based taxonomic classification, similarities between
Helicosporidium cells and trebouxiophyte green algae have also been highlighted at the cellular level, principally in regards to the vegetative cell multiplication and the autosporulation process [
8,
16].
The identification of
Helicosporidium spp. as non-photosynthetic green algae prompted several studies aimed at characterizing chloroplast-like genetic material, since achlorophytic algae and plants have been shown to retain vestigial chloroplasts, or plastids. Amplification and analysis of plastid 16S rDNA gene fragments suggested that
Helicosporidium cells had retained such an organelle [
29]. Sequencing of a cluster of protein encoding genes demonstrated that the
Helicosporidium plastid DNA was potentially transcribed [
30], and a subset of expressed sequences tags annotated as homologs to nucleus-encoded, plastid-targeted proteins offered additional indirect evidence for a plastid and its included genome [
31]. Although the organelle itself has yet to be observed in ultra-thin sections of
Helicosporidium cells, the presence of a vestigial chloroplast has been thoroughly demonstrated by the sequencing of the entire plastid genome [
32]. This genome lacks genes involved in photosynthesis and is so reduced that it is among the smallest known plastid genome. Overall, the plastid genome, and the phylogenetic inferred from its genes, have confirmed that
Helicosporidium spp. are highly specialized non-photosynthetic trebouxiophyte green algae. Additional phylogenomic evidence was most recently provided by the analysis of the complete mitochondrial genome for
Helicosporidium Sj-1 [
33].
Trebouxiophyte algae include
Coccomyxa spp., which are known pathogens of invertebrates such as mussels, scallops, geoducks, and starfishes [
34,
35,
36]. However,
Coccomyxa spp. are photosynthetic and have never been reported in insect hosts. The non-photosynthetic
Helicosporidium spp. remain the only described achlorophytic entomopathogenic algae, and recent phylogenetic analyses indicated that the genera
Helicosporidium and
Coccomyxa are not closely related [
37]. Virtually all phylogeny reconstructions have associated
Helicosporidium with
Prototheca, which harbors emerging pathogens of vertebrates, including isolates infecting humans, pets, and farm animals [
38]. Both
Prototheca and
Helicosporidium are non photosynthetic, and since most phylogenetic analyses have depicted
Helicosporidium and
Prototheca as members of a monophyletic clade, it is hypothesized that the loss of photosynthesis represents a synapomorphic character for these algae. Only selected species of
Prototheca are known to be pathogenic, and although there is similarities in cell division, pathogenic isolates of
P. wickerhamii or
P. zopfii have never been associated with a cyst stage, or filamentous cells, similar to
Helicosporidium. To date, these unique cellular structures are thought to be specific to the genus
Helicosporidium and, potentially, to its infectious process.
In addition to the studies aimed at precisely positioning
Helicosporidium within a taxonomic framework, independent phylogenetic analyses focused on the genus
Prototheca have incorporated the sequences generated from
Helicosporidium spp. and confirmed the close relationship between the two genera [
39,
40]. Both
Prototheca and
Helicosporidium have appeared in a monophyletic clade that included the photosynthetic species
Auxenochlorella protothecoides, and that has been referred to as the
Auxenochlorella-
Helicosporidium-
Prototheca (AHP) clade [
39]. In taxa-rich phylogenetic reconstructions focused on these genera, the species-level relationships have consistently been obscured by very poor resolution, and therefore the precise taxonomic position of
Helicosporidium within the Trebouxiophyceae remains unclear. Lineages within the class Trebouxiophyceae are currently poorly resolved [
41], and in the case of the AHP clade, it has been further complicated by the proposals for two novel
Prototheca species, which have yet to appear in large-scale taxonomic classifications [
42,
43]. A general consensus might be emerging, as the inclusion of
Helicosporidium has consistently led to phylogenetic trees that depicted
Prototheca as a paraphyletic genus. Recent studies indicated that
Helicosporidium is a sister taxon to a strongly supported clade that includes
P. zopfii,
P. ulmea,
P. moriformis,
P. stagnora, and the newly proposed
P. blaschkeae [
37,
40]. Other species, such as
P. wickerhamii and
P. cutis, were not identified as members of this
Prototheca sensu stricto clade, and therefore several authors have suggested that they should be re-assigned to alternative genera [
39,
40]. Although they have yet to include comparative analyses with
Helicosporidium, alternative cladistic approaches, based on nutritional requirements [
40], or biochemical profiles [
38,
39], have confirmed the strong heterogeneity of the genus
Prototheca. To date, the closest relative to
Helicosporidium spp. remain unidentified. A refined and potentially revised
Prototheca taxonomy may be necessary to precisely establish species-level relationships between non‑photosynthetic trebouxiophyte algae, and determine if the acquisition of pathogenicity occurred once or multiple times in these algae.
Attempts to obtain strongly supported phylogenetic trees featuring both
Helicosporidium and
Prototheca have led to the generation of sequence data from multiple
Helicosporidium isolates, and revealed an unsuspected genetic diversity. Sequences from plastid 16S rDNA genes [
29], nuclear 18S rDNA and β-tubulin genes [
36] contained polymorphic loci, suggesting that distinct
Helicosporidium isolates might be differentiated at a molecular level. In phylogenetic trees, all
Helicosporidium isolates were grouped in a strongly monophyletic clade, but analyses indicated that isolates collected from coleopteran hosts (strain Dm-1 and Cs-1) were more closely related to each other than they were to the dipteran isolate (strain Sj-1) [
36]. Combined with the previously noted differences in relation to morphological structures, and infectivity and virulence, these polymorphic characters may serve as a basis to develop comprehensive studies aimed at distinguishing more than one
Helicosporidium species. A refined understanding of the
Helicosporidium biodiversity may help establishing whether the currently observed host range for these pathogens is reflective of one or more species.
5. The Helicosporidium Host Range Includes a Wide Variety of Insects and Other Invertebrates
Insect infections with
Helicosporidium have been reported in three orders: Lepidoptera, Coleoptera and Diptera (
Table 1). Overall, 23 species (11 families and 16 genera) of insects are known natural hosts for these pathogenic algae. Coleopteran and dipteran infections are most common. Only two instances of lepidopteran infections have been reported. A hepialid larva was first identified as a
Helicosporidium host [
3]. Later,
Helicosporidium parasiticum was catalogued as a pathogen of the light brown apple moth (
Epiphyas postvittana) in New Zealand [
44]. In laboratory settings, lepidopteran larvae have been used extensively as heterologous hosts, clearly demonstrating that these insects are susceptible to infections [
8,
14,
15,
16,
17,
20,
21,
22,
23,
24].
Table 1.
Reported insect host record for Helicosporidium spp.
Table 1.
Reported insect host record for Helicosporidium spp.
| Original insect hosts | Location | Heterologous insect hosts 1 | Reference |
|---|
| Diptera | | | |
| Ceratopogonidae | | | |
| Dasyhelea obscura | England | nr | [1] |
| Rhiphidae | | | |
| Mycetobia pallipes | England | nr | [1] |
| Culicidae | | | |
| Culex territans | USA | nr | [45] |
| Culex nigripalpus | USA | Diptera (15) Coleoptera (3) Lepidoptera (2) | [26] |
| Culex quinquefasciatus | Thailand | nr | [24] |
| Culex pipiens | Egypt | Diptera (4) Lepidoptera (1) | [9] |
| Aedes aegypti | Thailand | Diptera (3) Lepidoptera (1) | [24] |
| Sciaridae | | | |
| Ctenosciara hyalipennis | Germany | nr | [46] |
| Simuliidae | | | |
| Simulium jonesi | USA | Diptera (4) Coleoptera (1) Lepidoptera (4) | [8] |
| Coleoptera | | | |
| Nitidulidae | | | |
| Carpophilus dimidiatus | USA | nr | [47] |
| Carpophilus freeman | USA | nr | [47] |
| Carpophilus hemipterus | USA | nr | [47] |
| Carpophilus mutilatus | USA, Mexico | Diptera (1) Coleoptera (9) Lepidoptera (5) | [47,48] |
| Carpophilus pilosellus | USA | nr | [47] |
| Conotelus stenoides | USA | nr | [47] |
| Stelidota geminata | USA | nr | [47] |
| Urophorus humeralis | Mexico | nr | [47] |
| Scarabaeidae | | | |
| Orycetes monoceros | Tanzania | nr | [49] |
| Curculionidae | | | |
| Cyrtobagous salviniae | South Africa | Diptera (3) Coleoptera (1) Lepidoptera (2) | [12] |
| Dendroctonus micans | Turkey | nr | [10] |
| Rhizophaginae | | | |
| Rhizophagus grandis | Turkey | nr | [11,13] |
| Lepidoptera | | | |
| Hepialidae | | | |
| Hepialis pallens | Argentina | nr | [3] |
| Tortricidae | | | |
| Epiphyas postvittana | New Zealand | nr | [44] |
Similar to the two infections reported in Lepidoptera, most coleopteran and dipteran accounts have corresponded to episodic observations (
Table 1). However, notable exceptions exist. In Diptera,
Helicosporidium infections have been recurrently reported in mosquitoes, especially in the genus
Culex, which has been found as a host on four different occasions on various continents (North America, Asia, Africa), suggesting that
Helicosporidium may be ubiquitous and chronic in mosquito populations [
9,
24,
26,
45]. Bioassays demonstrated that mosquito
Helicosporidium isolates were infectious to additional mosquito species [
9,
26]. A large survey of nitidulid beetles pathogens suggested that
Helicosporidium might be widespread in Coleoptera, as infections were reported in over 50% of the examined species [
47]. A tentative identification in the rhinoceros beetle [
49], combined with the recent detection of
Helicosporidium in two members of the Curculionidae family [
10,
12] supports the possibility that coleopterans serve as major hosts for these pathogens. The first study aimed at investigating the abundance and importance of
Helicosporidium on natural insect populations was recently performed on the coleopteran host
Dendroctonus micans, and results indicated that the pathogen is widely distributed and can be detected over a period of three years in different geographic locales, with estimated infection rates reaching 71% [
50]. A similar study focused on the occurrence of
Helicosporidium in a second coleopteran host, the predator beetle
Rhizophagus grandis (a predator of
D. micans), although it was limited to laboratory-reared insect [
51]. These two studies suggest that
Helicosporidium investigations may move from isolated and occasional reports of infection in a few insects to a broader understanding of the abundance and impact of these organisms in insect populations.
In addition to the observation that
Helicosporidium can be detected in Lepidoptera, Coleoptera and Diptera, one of the most striking and recurrent characteristics of these insect pathogens is arguably their ability to be horizontally transferred irrespectively of the host order from which they have been isolated. Isolates detected in dipteran hosts can readily infect coleopterans or lepidopterans [
8,
9,
16,
17,
20,
21,
22,
23]. Isolates collected in Coleoptera have showed similar broad host range, and were used to infect Lepidoptera or Diptera taxa [
12,
14,
15,
22,
23,
48]. The most comprehensive study of
Helicosporidium host range involved an isolate collected from the nitidulid beetle
Carpophilus mutilatus that was transmitted to nine other coleopteran species, five lepidopteran species and one dipteran species (Table 1) [
48]. This isolate also failed to infect orthopteran and hymenopteran insects [
48], providing the basis for, and supporting, the current understanding that the
Helicosporidium host range is restricted to three insect orders: Coleoptera, Lepidoptera and Diptera, as indicated in
Table 1. This study was also instrumental is demonstrating that the
Helicosporidium host range includes non-insect invertebrates, as the coleopteran isolate was shown to be infectious to three species of mites. Mites and collembolans have been reported as potential natural hosts for
Helicosporidium, as early as during the first description of
Helicosporidium parasiticum [
1,
5,
6,
17,
46]. In addition,
Helicosporidium spp. were detected in trematodes [
7] and cladocerans [
4]. Overall, pathosystems involving non-insect hosts remain largely uncharacterized.
Finally, in agreement with the observation that the cuticle of heavily infected insect hosts may rupture and release
Helicosporidium cysts and vegetative cells in the environment [
1,
52], several reports have indicated that
Helicosporidium can be isolated [
17,
25,
53], or at least detected [
37], from lentic water samples such as ponds or ditches. Pathogenicity of these isolates was evaluated in the corn earworm
Heliothis zea, or mosquitoes [
17,
25,
53], suggesting that pathogenic isolates can be uncovered independently of host infections. When tested on mosquitoes, a
Helicosporidium isolate collected from field water samples was compared to the Thailand mosquito isolate (from
Aedes aegyptii), and was shown to be not as effective or as lethal [
17]. This result prompted the proposal for distinct species of
Helicosporidium, and was reminiscent of other comparative studies that indicated that pathogenicity to
Culex pipiens quinquefasciatus was variable [
26]. Although a
Helicosporidium isolate from a coleopteran host was reportedly infectious to
Cx. p. quinquefasciatus [
48], this mosquito species appeared non-susceptible to an isolate collected from
Cx. nigripalpus [
26]. Although rare, the noted differences in host susceptibility and
Helicosporidium virulence suggest that the list of natural hosts presented in
Table 1 may be reflective of the combined range of several isolates that have yet to be formally distinguished and taxonomically separated.
As noted previously [
37], the ability to consistently detect
Helicosporidium in selected environments [
17,
25,
37,
53] or insect populations [
50,
51] will likely further the current understanding on these organisms by not only providing robust ecological data to estimate their impact on host populations but also expanding the current knowledge of
Helicosporidium biodiversity and the identification of polymorphic characters that may be informative for the distinction of several
Helicosporidium species.