PCR Diagnosis of Small Hive Beetles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Genomic DNA Extraction
2.3. Species-Specific PCR Assays
2.4. Bees as Matrices for PCR-Based Diagnostics
3. Results
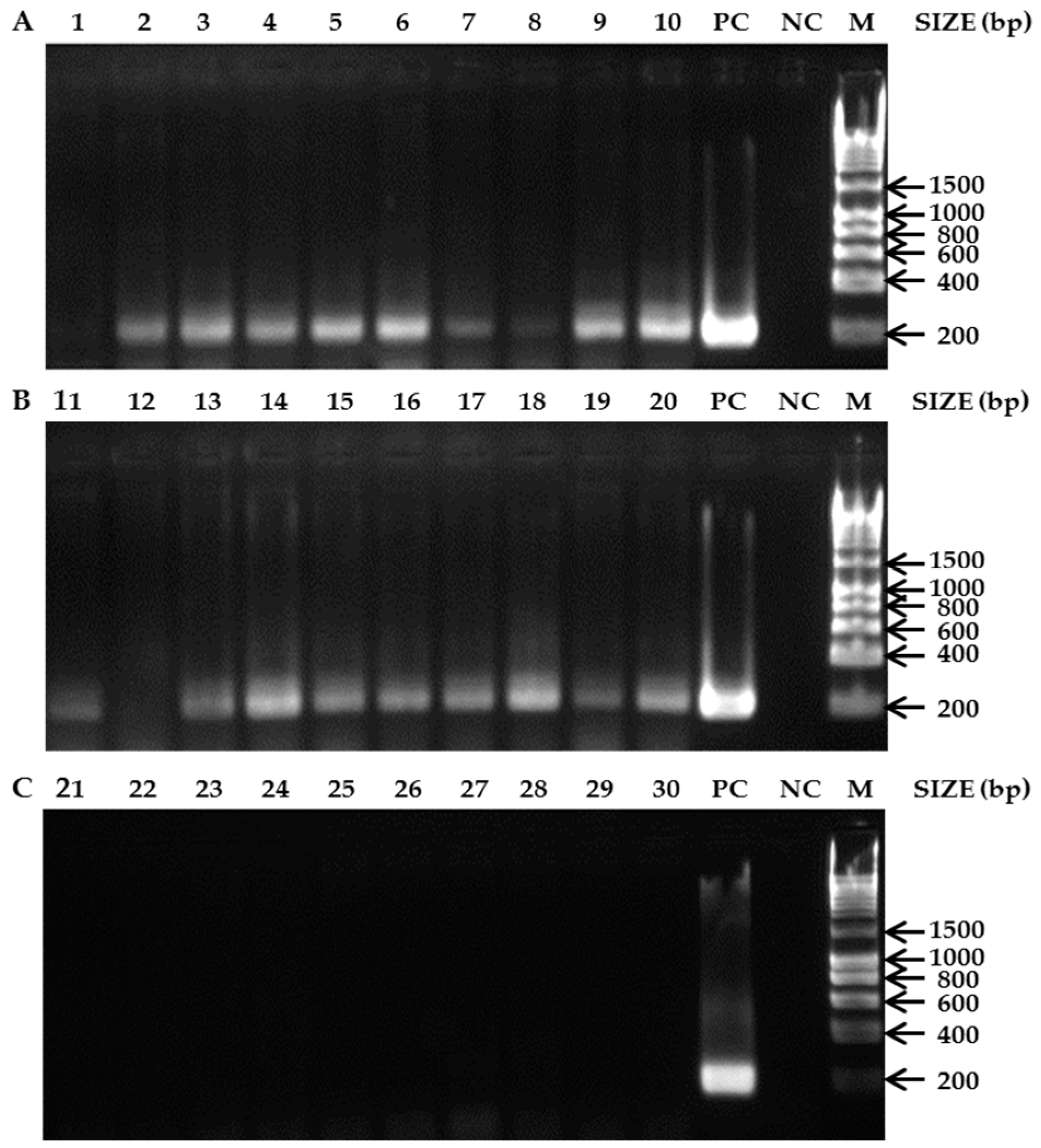
3.1. Species-Specific PCR Assays
3.2. Bees as Matrices for PCR-Based Diagnostics
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Neumann, P.; Pettis, J.S.; Schäfer, M.O. Quo vadis Aethina tumida? Biology and control of small hive beetles. Apidologie 2016, 47, 427–466. [Google Scholar] [CrossRef] [Green Version]
- Spiewok, S.; Neumann, P. Cryptic low-level reproduction of small hive beetles in honeybee colonies. J. Apic. Res. 2006, 45, 47–48. [Google Scholar] [CrossRef]
- Pirk, C.W.W.; Neumann, P. Small hive beetles are facultative predators of adult honey bees. J. Insect Behav. 2013, 26, 796–803. [Google Scholar] [CrossRef]
- Neumann, P.; Elzen, P.J. The biology of the small hive beetle (Aethina tumida, Murray): Gaps in our knowledge of an invasive species. Apidologie 2004, 35, 229–247. [Google Scholar] [CrossRef]
- Hepburn, H.R.; Radloff, S.E. Honeybees of Africa; Springer: Berlin, Germany, 1998. [Google Scholar]
- Toufailia, H.A.; Alves, D.A.; Bená, D.C.; Bento, J.M.S.; Iwanicki, N.S.A.; Cline, A.R.; Ellis, J.D.; Ratnieks, F.L.W. First record of small hive beetle, Aethina tumida Murray, in South America. J. Apic. Res. 2017, 56, 76–80. [Google Scholar] [CrossRef]
- Lee, S.; Hong, K.; Cho, Y.S.; Choi, Y.S.; Yoo, M.; Lee, S. Review of the subgenus Aethina Erichson s. str. (Coleoptera: Nitidulidae: Nitidulinae) in Korea, reporting recent invasion of small hive beetle, Aethina tumida. J. Asia Pac. Entomol. 2017, 20, 553–558. [Google Scholar] [CrossRef]
- Schäfer, M.O.; Cardaio, I.; Cilia, G.; Cornelissen, B.; Crailsheim, K.; Formato, G.; Akinwande, K.L.; Le Conte, Y.; Mutinelli, F.; Nanetti, A.; et al. How to slow the global spread of small hive beetles, Aethina tumida. Biol. Invasions 2018. under consideration. [Google Scholar]
- Evans, J.D.; Pettis, J.S.; Shimanuki, H. Mitochondrial DNA relationships in an emergent pest of honey bees: Aethina tumida (Coleoptera: Nitidulidae) from the United States and Africa. Ann. Entomol. Soc. Am. 2000, 93, 415–420. [Google Scholar] [CrossRef]
- Ward, L.; Brown, M.; Neumann, P.; Wilkins, S.; Pettis, J.; Boonham, N. A DNA method for screening hive debris for the presence of small hive beetle (Aethina tumida). Apidologie 2007, 38, 272–280. [Google Scholar] [CrossRef]
- Neumann, P.; Evans, J.D.; Pettis, J.S.; Pirk, C.W.W.; Schaefer, M.O.; Tanner, G.; Ellis, J.D. Standard methods for small hive beetle research. J. Apic. Res. 2013, 52, 1–32. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M. Primer3—New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Harpur, B.A.; Zayed, A. Accelerated evolution of innate immunity proteins in social insects: Adaptive evolution or relaxed constraint? Mol. Biol. Evol. 2013, 30, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; Bland, J.M. Statistics Notes: Diagnostic tests 1: Sensitivity and specificity. Br. Med. J. 1994, 308, 1552. [Google Scholar] [CrossRef]
- Neumann, P.; Moritz, R.F.A.; Mautz, D. Colony evaluation is not affected by the drifting of drone and worker honeybees (Apis mellifera carnica) at a performance testing apiary. Apidologie 2000, 31, 67–79. [Google Scholar] [CrossRef]
- Spiewok, S.; Duncan, M.; Spooner-Hart, R.; Pettis, J.S.; Neumann, P. Small hive beetle, Aethina tumida, populations II: Dispersal of small hive beetles. Apidologie 2008, 39, 683–693. [Google Scholar] [CrossRef]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouessou Idrissou, F.; Huang, Q.; Yañez, O.; Akinwande, K.L.; Neumann, P. PCR Diagnosis of Small Hive Beetles. Insects 2018, 9, 24. https://doi.org/10.3390/insects9010024
Ouessou Idrissou F, Huang Q, Yañez O, Akinwande KL, Neumann P. PCR Diagnosis of Small Hive Beetles. Insects. 2018; 9(1):24. https://doi.org/10.3390/insects9010024
Chicago/Turabian StyleOuessou Idrissou, Franck, Qiang Huang, Orlando Yañez, Kayode Lawrence Akinwande, and Peter Neumann. 2018. "PCR Diagnosis of Small Hive Beetles" Insects 9, no. 1: 24. https://doi.org/10.3390/insects9010024






