Epigenetic Regulation of a Heat-Activated Retrotransposon in Cruciferous Vegetables
Abstract
:1. Introduction
2. Results
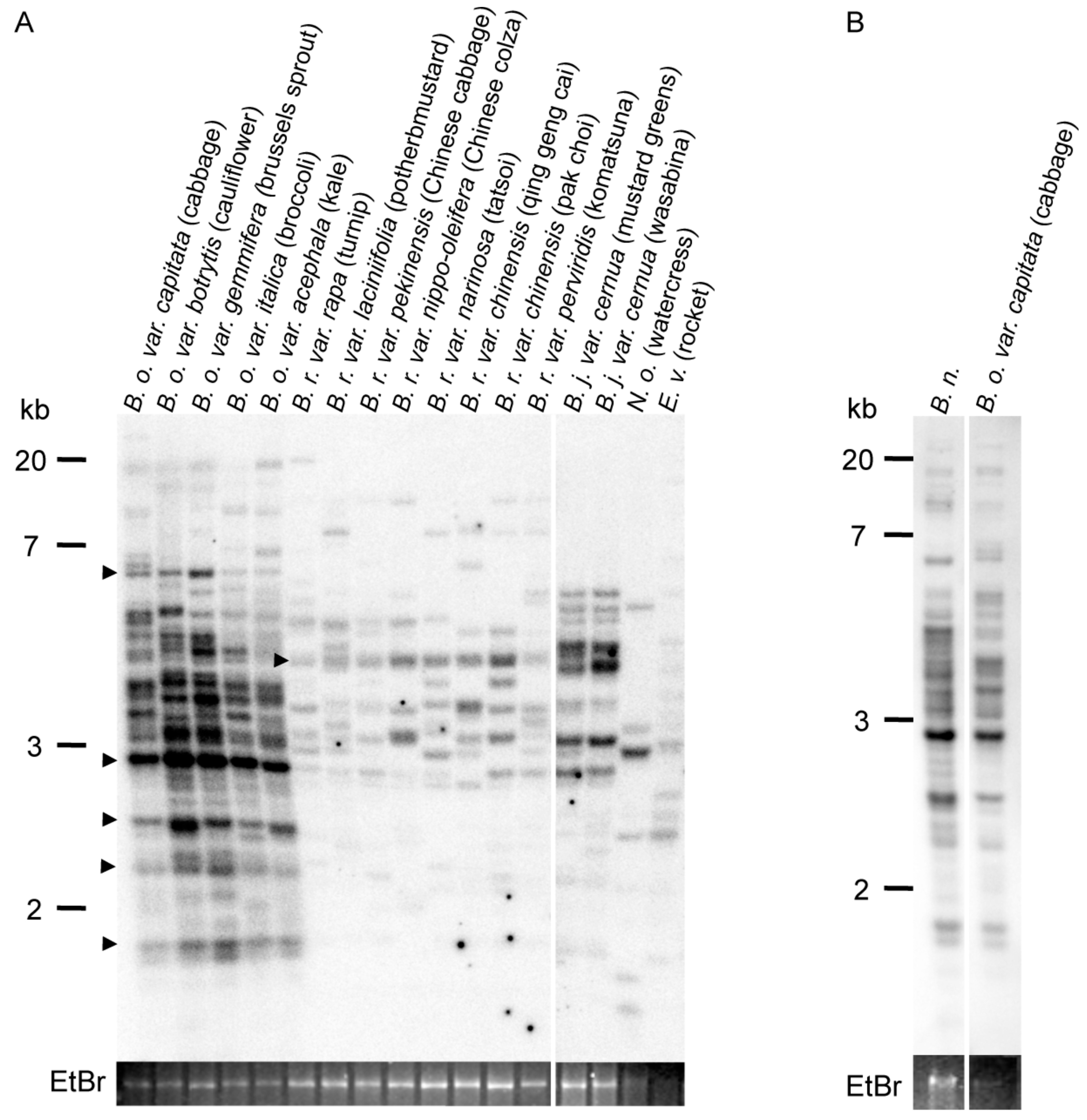
2.1. Copy Number of ONSEN-Like Elements
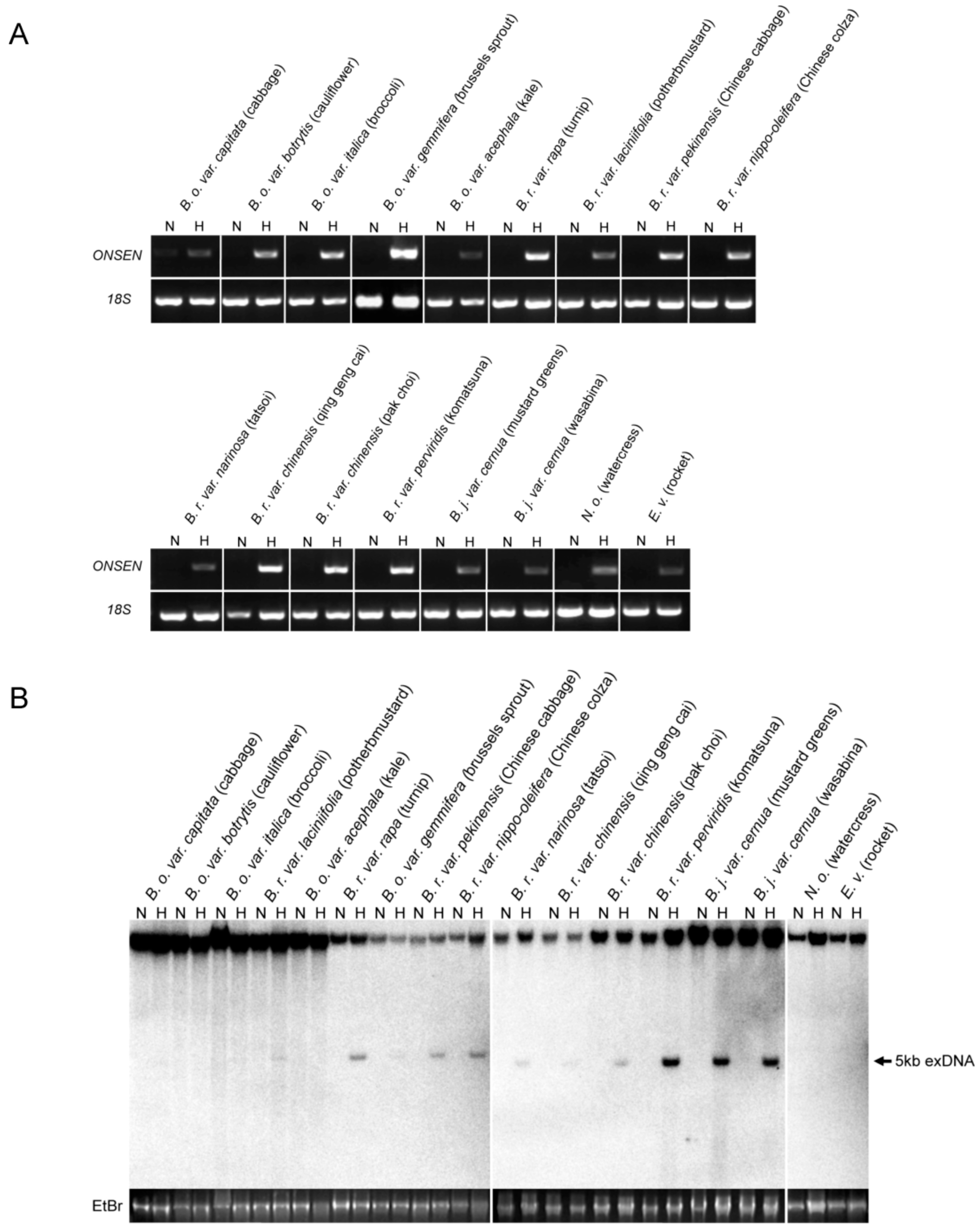
2.2. Heat-Activation of OLE in Cruciferous Vegetables
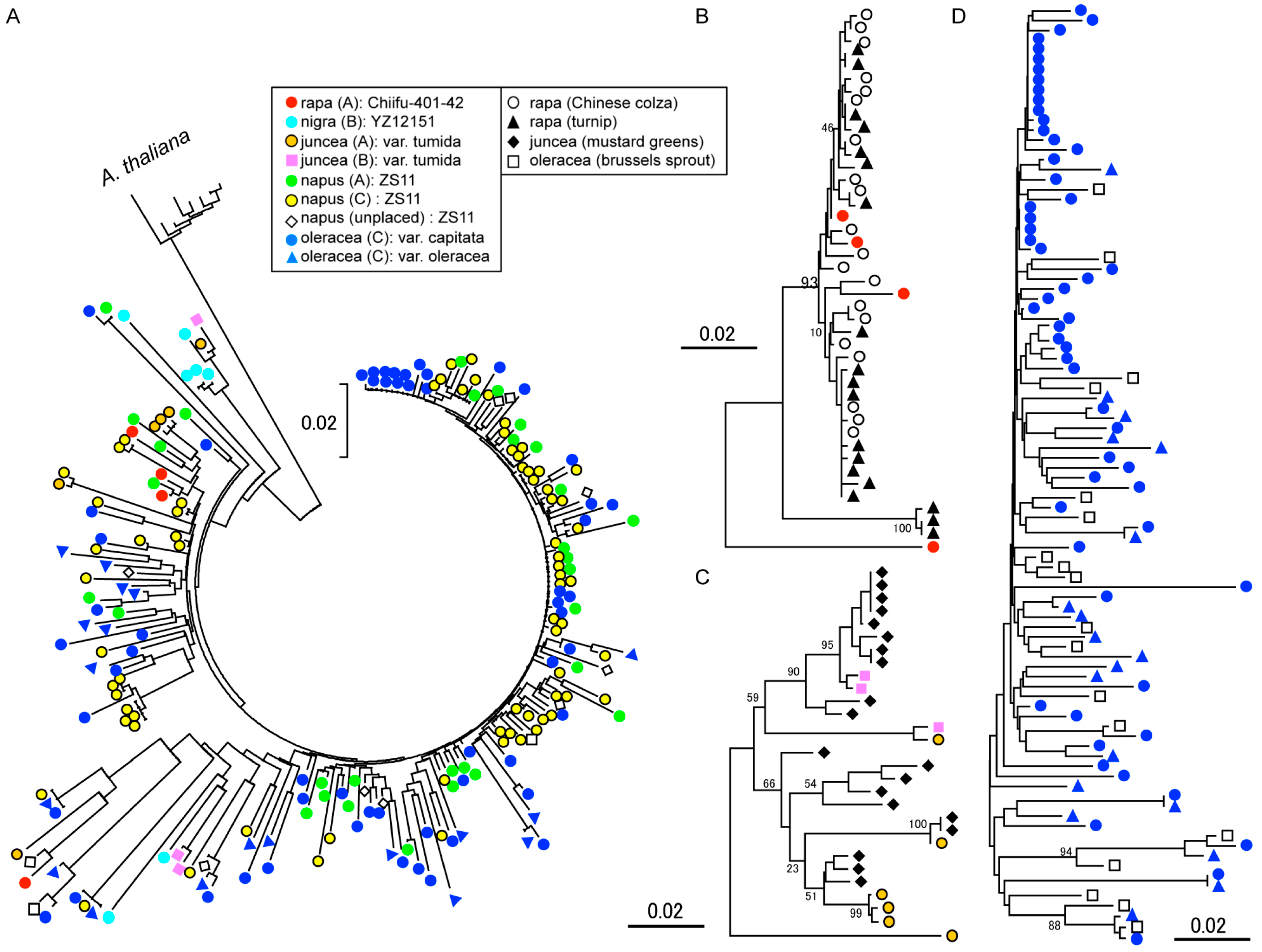
2.3. Phylogenic Analysis of OLEs in Cruciferous Vegetables
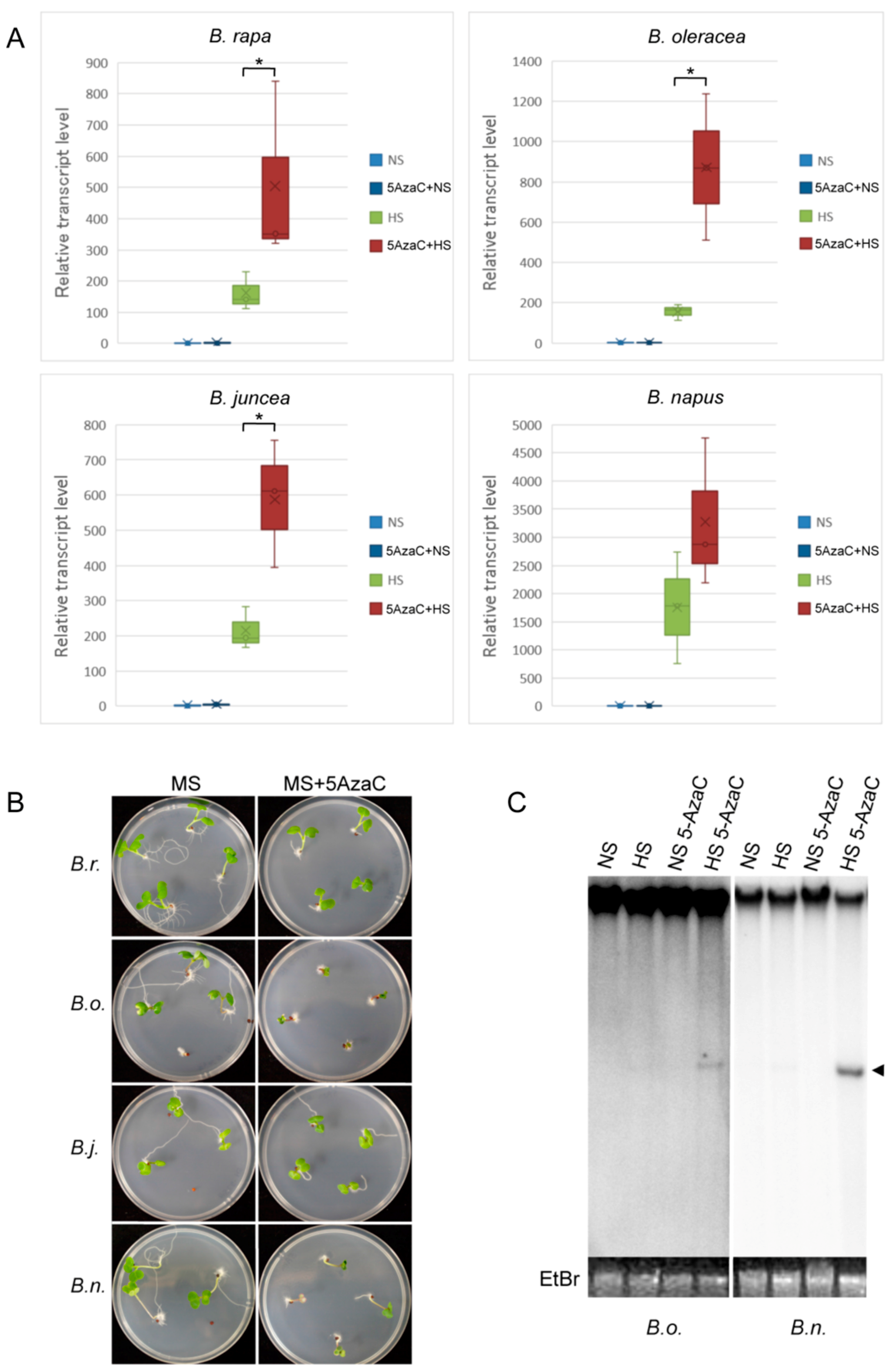
2.4. Epigenetic Regulation of OLE in Cruciferous Vegetables
3. Discussion
4. Materials and Methods
4.1. Plant Material and Stress Treatments
4.2. Southern Blot Analysis
4.3. RT-PCR
4.4. Sequence Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kumar, A.; Bennetzen, J.L. Plant retrotransposons. Annu. Rev. Genet. 1999, 33, 479–532. [Google Scholar] [CrossRef] [PubMed]
- SanMiguel, P.; Gaut, B.S.; Tikhonov, A.; Nakajima, Y.; Bennetzen, J.L. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998, 20, 43–45. [Google Scholar] [CrossRef] [PubMed]
- Vitte, C.; Bennetzen, J.L. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 2006, 103, 17638–17643. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Jacobsen, S.E. Epigenetic inheritance in plants. Nature 2007, 447, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. Epigenetic Regulation of Transposable Elements in Plants. Annu. Rev. Plant Biol. 2009, 60, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Jiang, N.; Wessler, S.R. Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 2002, 3, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Gaubert, H.; Bucher, E.; Mirouze, M.; Vaillant, I.; Paszkowski, J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 2011, 472, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Cavrak, V.V.; Lettner, N.; Jamge, S.; Kosarewicz, A.; Bayer, L.M.; Scheid, O.M. How a Retrotransposon Exploits the Plant's Heat Stress Response for Its Activation. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Masuta, Y.; Nozawa, K.; Takagi, H.; Yaegashi, H.; Tanaka, K.; Ito, T.; Saito, H.; Kobayashi, H.; Matsunaga, W.; Masuda, S.; et al. Inducible Transposition of a Heat-Activated Retrotransposon in Tissue Culture. Plant Cell Physiol. 2017, 58, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Kim, J.M.; Matsunaga, W.; Saze, H.; Matsui, A.; Endo, T.A.; Harukawa, Y.; Takagi, H.; Yaegashi, H.; Masuta, Y.; et al. A Stress-Activated Transposon in Arabidopsis Induces Transgenerational Abscisic Acid Insensitivity. Sci. Rep. 2016, 6, 23181. [Google Scholar] [CrossRef] [PubMed]
- Warwick, S.I.; Francis, A.; Al-Shehbaz, I.A. Brassicaceae: Species checklist and database on CD-Rom. Plant Syst. Evol. 2006, 259, 249–258. [Google Scholar] [CrossRef]
- Maumus, F.; Allen, A.E.; Mhiri, C.; Hu, H.H.; Jabbari, K.; Vardi, A.; Grandbastien, M.A.; Bowler, C. Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genom. 2009, 10, 624. [Google Scholar] [CrossRef] [PubMed]
- Naito, K.; Zhang, F.; Tsukiyama, T.; Saito, H.; Hancock, C.N.; Richardson, A.O.; Okumoto, Y.; Tanisaka, T.; Wessler, S.R. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 2009, 461, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons Control Fruit-Specific, Cold-Dependent Accumulation of Anthocyanins in Blood Oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed]
- Makarevitch, I.; Waters, A.J.; West, P.T.; Stitzer, M.; Hirsch, C.N.; Ross-Ibarra, J.; Springer, N.M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015, 11, e1004915. [Google Scholar] [CrossRef]
- Ito, H.; Yoshida, T.; Tsukahara, S.; Kawabe, A. Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene 2013, 518, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Pietzenuk, B.; Markus, C.; Gaubert, H.; Bagwan, N.; Merotto, A.; Bucher, E.; Pecinka, A. Recurrent evolution of heat-responsiveness in Brassicaceae COPIA elements. Genome Biol. 2016, 17, 209. [Google Scholar] [CrossRef] [PubMed]
- Mari-Ordonez, A.; Marchais, A.; Etcheverry, M.; Martin, A.; Colot, V.; Voinnet, O. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 2013, 45, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Grandbastien, M.A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998, 3, 181–187. [Google Scholar] [CrossRef]
- Liu, B.; Wendel, J.F. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 2000, 43, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.P.; Si, Y.; Hanson, R.E.; Crane, C.F.; Price, H.J.; Stelly, D.M.; Wendel, J.F.; Paterson, A.H. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Res. 1998, 8, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Miura, A.; Kato, M.; Watanabe, K.; Kawabe, A.; Kotani, H.; Kakutani, T. Genomic localization of endogenous mobile CACTA family transposons in natural variants of Arabidopsis thaliana. Mol. Genet. Genom. 2004, 270, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Church, G.M.; Gilbert, W. Genomic sequencing. Proc. Natl. Acad. Sci. USA 1984, 81, 1991–1995. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Wang, H.Z.; Wang, J.; Sun, R.F.; Wu, J.; Liu, S.Y.; Bai, Y.Q.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Parkin, I.A.; Koh, C.; Tang, H.; Robinson, S.J.; Kagale, S.; Clarke, W.E.; Town, C.D.; Nixon, J.; Krishnakumar, V.; Bidwell, S.L.; et al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014, 15, R77. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nozawa, K.; Kawagishi, Y.; Kawabe, A.; Sato, M.; Masuta, Y.; Kato, A.; Ito, H. Epigenetic Regulation of a Heat-Activated Retrotransposon in Cruciferous Vegetables. Epigenomes 2017, 1, 7. https://doi.org/10.3390/epigenomes1010007
Nozawa K, Kawagishi Y, Kawabe A, Sato M, Masuta Y, Kato A, Ito H. Epigenetic Regulation of a Heat-Activated Retrotransposon in Cruciferous Vegetables. Epigenomes. 2017; 1(1):7. https://doi.org/10.3390/epigenomes1010007
Chicago/Turabian StyleNozawa, Kosuke, Yuki Kawagishi, Akira Kawabe, Mio Sato, Yukari Masuta, Atsushi Kato, and Hidetaka Ito. 2017. "Epigenetic Regulation of a Heat-Activated Retrotransposon in Cruciferous Vegetables" Epigenomes 1, no. 1: 7. https://doi.org/10.3390/epigenomes1010007





