EZH2 Single Nucleotide Variants (SNVs): Diagnostic and Prognostic Role in 10 Solid Tumor Types
Abstract
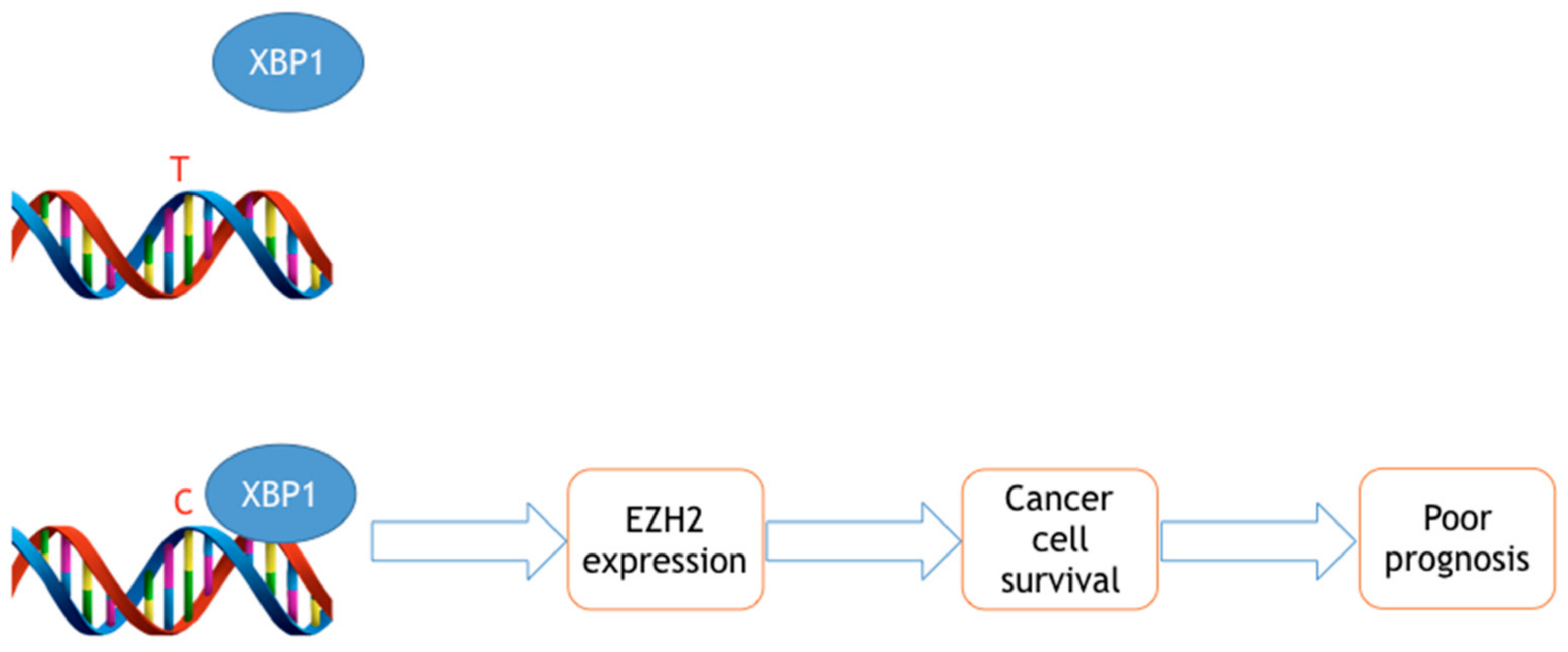
:1. Introduction
2. EZH2 SNVs Associated with Cancer Susceptibility
3. EZH2 SNVs Associated with Prognosis and Response to Therapy
4. Conclusions
- 1)
- 2)
- Functional studies on the role of clinically relevant SNVs, identified in step 1. These studies should be conducted at least in vitro and in human tissues, to confirm the effects of each variant upon EZH2 expression or other function.
Author Contributions
Conflicts of Interest
References
- Aloia, L.; Di Stefano, B.; Di Croce, L. Polycomb Complexes in Stem Cells and Embryonic Development. Development 2013, 140, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.; Mignon, C.; Hetet, G.; Grandchamps, B.; Fontes, M.; Colleaux, L. The Human EZH2 Gene: Genomic Organisation and Revised Mapping in 7q35 within the Critical Region for Malignant Myeloid Disorders. Eur. J. Hum. Genet. 2000, 8, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Mozgova, I.; Hennig, L. The Polycomb Group Protein Regulatory Network. Annu. Rev. Plant Biol. 2015, 66, 269–296. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Sauvageau, G. Polycomb Group Proteins: Multi-Faceted Regulators of Somatic Stem Cells and Cancer. Cell Stem Cell 2010, 7, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Borbone, E.; Troncone, G.; Ferraro, A.; Jasencakova, Z.; Stojic, L.; Esposito, F.; Hornig, N.; Fusco, A.; Orlando, V. Enhancer of Zeste Homolog 2 Overexpression has a Role in the Development of Anaplastic Thyroid Carcinomas. J. Clin. Endocrinol. Metab. 2011, 96, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.; Otte, A.P.; Hayes, D.F.; et al. Chinnaiyan, A.M. EZH2 is a Marker of Aggressive Breast Cancer and Promotes Neoplastic Transformation of Breast Epithelial Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Gu, L.; Cao, Y.; Fan, X.; Zhang, F.; Sang, M. Aberrant Overexpression of EZH2 and H3K27me3 Serves as Poor Prognostic Biomarker for Esophageal Squamous Cell Carcinoma Patients. Biomarkers 2016, 21, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.C.; Yang, Z.L. Overexpression of EZH2 and Loss of Expression of PTEN is Associated with Invasion, Metastasis, and Poor Progression of Gallbladder Adenocarcinoma. Pathol. Res. Pract. 2011, 207, 472–478. [Google Scholar] [CrossRef] [PubMed]
- He, L.J.; Cai, M.Y.; Xu, G.L.; Li, J.J.; Weng, Z.J.; Xu, D.Z.; Luo, G.Y.; Zhu, S.L.; Xie, D. Prognostic Significance of Overexpression of EZH2 and H3k27me3 Proteins in Gastric Cancer. Asian Pac. J. Cancer Prev. 2012, 13, 3173–3178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, X.; Chen, L.; Zhang, Z.; Feng, S. EZH2 Overexpression is Associated with Poor Prognosis in Patients with Glioma. Oncotarget. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Shen, J.; Gao, Y.; Zhou, Y.; Yu, Z.; Hornicek, F.; Kan, Q.; Duan, Z. Overexpression of EZH2 is Associated with the Poor Prognosis in Osteosarcoma and Function Analysis Indicates a Therapeutic Potential. Oncotarget 2016, 7, 38333–38346. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yu, L.; Li, Z.; Shen, Y.; Wang, J.; Cai, J.; Xiao, L.; Wang, Z. Overexpression of EZH2 Contributes to Acquired Cisplatin Resistance in Ovarian Cancer Cells in Vitro and in Vivo. Cancer Biol. Ther. 2010, 10, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Melling, N.; Thomsen, E.; Tsourlakis, M.C.; Kluth, M.; Hube-Magg, C.; Minner, S.; Koop, C.; Graefen, M.; Heinzer, H.; Wittmer, C.; et al. Overexpression of Enhancer of Zeste Homolog 2 (EZH2) Characterizes an Aggressive Subset of Prostate Cancers and Predicts Patient Prognosis Independently from pre- and Postoperatively Assessed Clinicopathological Parameters. Carcinogenesis 2015, 36, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Geng, H.; Qi, C.; Liu, Y.; Yue, D. Overexpression of YB1 and EZH2 are Associated with Cancer Metastasis and Poor Prognosis in Renal Cell Carcinomas. Tumor Biol. 2015, 36, 7159–7166. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Kaneda, A.; Tsuji, S.; Isagawa, T.; Yamamoto, S.; Fujita, T.; Yamanaka, R.; Tanaka, Y.; Nukiwa, T.; Marquez, V.E.; et al. PRC2 Overexpression and PRC2-Target Gene Repression Relating to Poorer Prognosis in Small Cell Lung Cancer. Sci. Rep. 2013, 3, 1911. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Jiang, Y.; Ma, S.; Chang, H.; Yi, C.; Cao, H.; Gao, Y.; Guo, H.; Hou, J.; Yan, J.; Sheng, Y.; Ren, X. EZH2 Promotes Invasion and Metastasis of Laryngeal Squamous Cells Carcinoma via Epithelial-Mesenchymal Transition Through H3K27me3. Biochem. Biophys. Res. Commun. 2016, 479, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, H.; Zhao, J.; Vieth, E.; Nephew, K.P.; Matei, D. EZH2 Inhibition Promotes Epithelial-to-Mesenchymal Transition in Ovarian Cancer Cells. Oncotarget 2016, 7, 84453–84467. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.W.; Gwak, S.Y.; Shim, G.A.; Liu, L.; Lim, Y.C.; Kim, J.M.; Jung, M.G.; Koo, B.S. EZH2 is Associated with Poor Prognosis in Head-and-Neck Squamous Cell Carcinoma via Regulating the Epithelial-to-Mesenchymal Transition and Chemosensitivity. Oral Oncol. 2016, 52, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Yu, J.; Dhanasekaran, S.M.; Kim, J.H.; Mani, R.S.; Tomlins, S.A.; Mehra, R.; Laxman, B.; Cao, X.; Yu, J.; et al. Repression of E-Cadherin by the Polycomb Group Protein EZH2 in Cancer. Oncogene 2008, 27, 7274–7284. [Google Scholar] [CrossRef] [PubMed]
- Herranz, N.; Pasini, D.; Díaz, V.M.; Francí, C.; Gutierrez, A.; Dave, N.; Escrivà, M.; Hernandez-Muñoz, I.; Di Croce, L.; Helin, K.; et al. Polycomb Complex 2 is Required for E-Cadherin Repression by the Snail1 Transcription Factor. Mol. Cell. Biol. 2008, 28, 4772–4781. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Sun, L.; Pikor, L.; Frumento, P.; Lam, W.L.; Helgason, C.D. Mutational Analysis of Polycomb Genes in Solid Tumours Identifies PHC3 Amplification as a Possible Cancer-Driving Genetic Alteration. Br. J. Cancer 2013, 109, 1699–1702. [Google Scholar] [CrossRef] [PubMed]
- Grzenda, A.; Lomberk, G.; Svingen, P.; Mathison, A.; Calvo, E.; Iovanna, J.; Xiong, Y.; Faubion, W.; Urrutia, R. Functional Characterization of EZH2β Reveals the Increased Complexity of EZH2 Isoforms Involved in the Regulation of Mammalian Gene Expression. Epigenetics Chromatin 2013, 6, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seim, N.B.; Kang, S.Y.; Bhandari, M.; Jones, R.G.; Teknos, T.N. Personalized Medicine Approach for an Exceptional Response to Multiple-recurrent and Metastatic HER2-Positive Oropharyngeal Squamous Cell Carcinoma. Ann. Otol. Rhinol. Laryngol. 2017, 126, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Su, K.J.; Lin, C.W.; Chen, M.K.; Yang, S.F.; Yu, Y.L. Effects of EZH2 Promoter Polymorphisms and Methylation Status on Oral Squamous Cell Carcinoma Susceptibility and Pathology. Am. J. Cancer Res. 2015, 5, 3475–3484. [Google Scholar] [PubMed]
- Yoon, K.A.; Gil, H.J.; Han, J.; Park, J.; Lee, J.S. Genetic Polymorphisms in the Polycomb Group Gene EZH2 and the Risk of Lung Cancer. J. Thorac. Oncol. 2010, 5, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.L.; Su, K.J.; Hsieh, Y.H.; Lee, H.L.; Chen, T.Y.; Hsiao, P.C.; Yang, S.F. Effects of EZH2 Polymorphisms on Susceptibility to and Pathological Development of Hepatocellular Carcinoma. PLoS ONE 2013, 8, e74870. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.L.; Su, K.J.; Hsieh, M.J.; Wang, S.S.; Wang, P.H.; Weng, W.C.; Yang, S.F. Impact of EZH2 Polymorphisms on Urothelial Cell Carcinoma Susceptibility and Clinicopathologic Features. PLoS ONE 2014, 9, e93635. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.B.; Guo, G.H.; Niu, Q.; Shi, N. Role of EZH2 Polymorphisms in Esophageal Squamous Cell Carcinoma Risk in Han Chinese Population. Int. J. Mol. Sci. 2014, 15, 12688–12697. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.B.; Sun, S.L.; Zheng, Q.L.; Zhang, L.; Zhu, Y.; Jin, G.H.; Xue, L.X. Genetic Alteration and Misexpression of Polycomb Group Genes in Hepatocellular Carcinoma. Am. J. Cancer Res. 2015, 5, 2969–2979. [Google Scholar] [PubMed]
- Chang, W.S.; Liao, C.H.; Tsai, C.W.; Hu, P.S.; Wu, H.C.; Hsu, S.W.; Hsiao, C.L.; Hsu, C.H.; Hung, Y.W.; Bau, D.T. Association of Enhancer of Zeste 2 (EZH2) Genotypes with Bladder Cancer Risk in Taiwan. Anticancer Res. 2016, 36, 4509–4514. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Du, W.D.; Wu, Q.; Liu, Y.; Chen, G.; Ruan, J.; Xu, S.; Yang, F.; Zhou, F.S.; Tang, X.F.; et al. EZH2 Genetic Variants Affect Risk of Gastric Cancer in the Chinese Han Population. Mol. Carcinog. 2014, 53, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Fornaro, L.; Paolicchi, E.; Masi, G.; Frumento, P.; Loupakis, F.; Salvatore, L.; Cremolini, C.; Schirripa, M.; Graziano, F.; et al. An EZH2 Polymorphism is Associated with Clinical Outcome in Metastatic Colorectal Cancer Patients. Ann. Oncol. 2012, 23, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Fornaro, L.; Crea, F.; Masi, G.; Paolicchi, E.; Loupakis, F.; Graziano, F.; Salvatore, L.; Ronzoni, M.; Ricci, V.; Cremolini, C.; et al. EZH2 Polymorphism and Benefit from Bevacizumab in Colorectal Cancer: Another Piece to the Puzzle. Ann. Oncol. 2012, 23, 1370–1371. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Fujisawa, J.; Yoshida, M. Identificational Activation Domain of TREB5, a CREB/ATF Family Protein that Binds to HTLV-1 Enhancer. J. Biochem. 1995, 117, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Fornaro, L.; Faviana, P.; De Gregorio, V.; Vivaldi, C.; Paolicchi, E.; Masi, G.; Loupakis, F.; Sensi, E.; Lupi, C.; Fontanini, G.; et al. Molecular and Pathological Characterization of the EZH2 rs3757441 Single Nucleotide Polymorphism in Colorectal Cancer. BMC Cancer 2015, 15, 874. [Google Scholar] [CrossRef] [PubMed]
- Paolicchi, E.; Pacetti, P.; Giovannetti, E.; Mambrini, A.; Orlandi, M.; Crea, F.; Romani, A.A.; Tartarini, R.; Danesi, R.; Peters, G.J.; et al. A Single Nucleotide Polymorphism in EZH2 Predicts overall Survival Rate in Patients with Cholangiocarcinoma. Oncol. Lett. 2013, 6, 1487–1491. [Google Scholar] [CrossRef] [PubMed]
- Tiffen, J.; Wilson, S.; Gallagher, S.J.; Hersey, P.; Filipp, F.V. Somatic Copy Number Amplification and Hyperactivating Somatic Mutations of EZH2 Correlate with DNA Methylation and Drive Epigenetic Silencing of Genes Involved in Tumor Suppression and Immune Responses in Melanoma. Neoplasia 2016, 18, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, F.; Shields, B.; Makhoul, I.; Hutchins, L.F.; Shalin, S.C.; Tackett, A.J. Role of EZH2 Histone Methyltransferase in Melanoma Progression and Metastasis. Cancer Biol. Ther. 2016, 17, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Fornaro, L.; Bocci, G.; Sun, L.; Farrar, W.L.; Falcone, A.; Danesi, R. EZH2 Inhibition: Targeting the Crossroad of Tumor Invasion and Angiogenesis. Cancer Metastasis Rev. 2012, 31, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, R.G.; Gehling, V.S.; Dakin, L.A.; Cook, A.S.; Nasveschuk, C.G.; Duplessis, M.; Iyer, P.; Balasubramanian, S.; Zhao, F.; Good, A.C.; et al. Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas. J. Med. Chem. 2016, 59, 9928–9941. [Google Scholar] [PubMed]
| SNV | Type of Variant | Risk Allele | Protective Allele | Molecular Function | Cancer | OR | CI 95% | p Value | Ref |
|---|---|---|---|---|---|---|---|---|---|
| rs12670401 | T > C, intron | C | Gastric cancer | 1.327 | 1.075–1.683 | 0.009 | [31] | ||
| rs2072407 | C > T, intron | n | TCheterozygote | Gastric cancer | 0.787 | 0.633–0.981 | 0.033 | [31] | |
| rs6464926 | C > T, intron | T | Gastric cancer | 1.310 | 1.059–1.619 | 0.012 | [31] | ||
| rs734004 | G > C, intron | CG heterozygote | Gastric cancer | 0.803 | 0.645–0.999 | 0.048 | [31] | ||
| rs734005 | T > C, intron | TC heterozygote | Gastric cancer | 0.799 | 0.642–0.995 | 0.045 | [31] | ||
| rs2302427 | G > C, exon, missense (D185H) | G | Concomitant increased expression of CBX8 and BMI1 may confer risk. PRC1 with PRC2 expression may be protective | HCC | 1.636 | 1.074–2.491 | 0.021 | [29] | |
| GG homozygote | 0.388 | 0.168–1.895 | 0.038 | [27] | |||||
| rs6950683 | T > C, 5’UTR | C | Hypermethylation of EZH2 in CC genotype patients compared to the TC genotype | OSCC | 0.791 | 0.66–0.948 | 0.011 | [24] | |
| C | HCC | 0.288 | 0.13–0.638 | <0.05 | [26] | ||||
| - | Lung cancer | 0.71 | 0.55–0.91 | 0.007 | [25] | ||||
| TC + CC genotypes | UCC | 0.565 | 0.382–0.835 | 0.004 | [27] | ||||
| rs3757441 | T > C, intron | C | OSCC | 0.820 | (0.683–0.984) | 0.033 | [24] | ||
| C | HCC | 0.273 | 0.116–0.645 | <0.05 | [26] | ||||
| - | Lung cancer | 0.73 | 0.57–0.94 | 0.015 | [25] | ||||
| C | EZH2 and H3K27me3 up-regulation | CRC | 0.009 | [32,35] | |||||
| rs887569 | C > T, intron | TT homozygote | Bladder cancer | - | - | 0.0146 | [30] |
| SNV | Location | Risk Allele | Protective Allele | Clinical Manifestation | Cancer | CI 95% | p Value | Ref |
|---|---|---|---|---|---|---|---|---|
| rs2302427 | transcript variant | G | Increase in invasive tumor stage | UCC | 0.220–0.794 | N/A | [27] | |
| rs3757441 | intron | C | T | Higher lymph–node-metastasis risk but a lower liver-cirrhosis risk | HCC | 1.747–208.155 | N/A | [26] |
| C | T | Shorter PFS and OS | mCRC | N/A | < 0.01 < 0.05 | [33] | ||
| C | T | Increased risk of tumor size, differentiation, T stage, and pathological stage | ESCC | N/A | 0.006 | [28] | ||
| rs6950683 | 5’UTR | C | T | Higher lymph–node-metastasis risk but a lower liver-cirrhosis risk | HCC | 1.733–208.866 | N/A | [26] |
| rs887569 | intron | C | T | Longer OS, reduced risk of mortality | Gallbladder | 0.33–1.05 | 0.026 | [36] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paolicchi, E.; Fornaro, L.; Landi, S.; Rigas, S.; Crea, F. EZH2 Single Nucleotide Variants (SNVs): Diagnostic and Prognostic Role in 10 Solid Tumor Types. Epigenomes 2017, 1, 18. https://doi.org/10.3390/epigenomes1030018
Paolicchi E, Fornaro L, Landi S, Rigas S, Crea F. EZH2 Single Nucleotide Variants (SNVs): Diagnostic and Prognostic Role in 10 Solid Tumor Types. Epigenomes. 2017; 1(3):18. https://doi.org/10.3390/epigenomes1030018
Chicago/Turabian StylePaolicchi, Elisa, Lorenzo Fornaro, Stefano Landi, Sushilaben Rigas, and Francesco Crea. 2017. "EZH2 Single Nucleotide Variants (SNVs): Diagnostic and Prognostic Role in 10 Solid Tumor Types" Epigenomes 1, no. 3: 18. https://doi.org/10.3390/epigenomes1030018






