Interaction and Binding Modes of bis-Ruthenium(II) Complex to Synthetic DNAs
Abstract
:1. Introduction
2. Experimental Section
3. Results
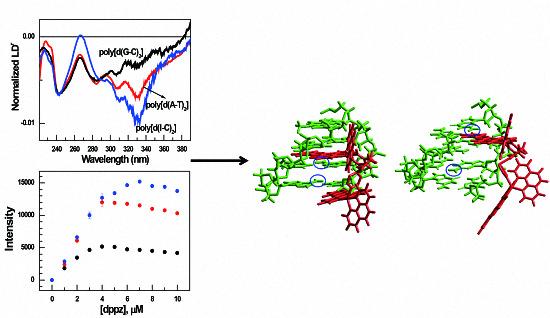
3.1. Absorption and Circular Dichroism
3.2. LDr Spectra
3.3. Luminescence and Quenching Measurement
4. Discussion
Binding Properties of Bis-Ru-Bpp Complex Bound to Polynucleotides
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PEG | PolyethyleneGlycole |
| CD | Circular dichrosim |
| LD | Linear dichroism |
References
- Kastan, M.B.; Bartek, J. Cell-cycle checkpoints and cancer. Nature 2004, 432, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Erkkila, K.E.; Odem, D.T.; Barton, J.K. Recognition and reaction of metallointercalators with DNA. Chem. Rev. 1999, 99, 2777–2796. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, C.; Thomas, J.A. Kinetically inert transition metal complexes that reversibly bind to DNA. Chem. Soc. Rev. 2003, 23, 215–524. [Google Scholar] [CrossRef]
- Xiong, Y.; Ji, L.N. Synthesis, DNA binding and DNA mediated luminescence quenching of Ru(II)polypyridine complexes. Coord. Chem. Rev. 1999, 185, 711–733. [Google Scholar] [CrossRef]
- Blasius, R.; Moucheron, C.; Mesmaeker, A.K.-D. Photoadducts of Metallic Compounds with Nucleic Acids—Role Played by the Photoelectron Transfer Process and by the TAP and HAT Ligands in the RuII Complexes. Eur. J. Inorg. Chem. 2004, 20, 3971–3979. [Google Scholar] [CrossRef]
- Nordén, B.; Lincoln, P.; Akerman, B.; Tuite, E. Metal Ions in Biological System; Sigel, A., Sigel, S., Eds.; Marcel Dekker: New York, NY, USA, 1996; Volume 33, pp. 177–252. [Google Scholar]
- Delaney, S.; Yoo, J.; Stemp, E.D.A.; Barton, J.K. Charge equilibration between two distinct sites in double helical DNA. Proc. Natl. Acad. Sci. USA 2004, 101, 10511–10516. [Google Scholar] [CrossRef] [PubMed]
- Brabec, V.; Kasparkova, J. Modifications of DNA by platinum complexes: Relation to resistance of tumors to platinum antitumor drugs. Drug Resist. Updates 2005, 8, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Hannon, M.J. Metal-based anticancer drugs: From a past anchored in platinum chemistry to a post-genomic future of diverse chemistry and biology. Pure Appl. Chem. 2007, 79, 2243–2261. [Google Scholar] [CrossRef]
- Clarke, M.; Zhu, F.; Frasca, D.R. Non-Platinum Chemotherapeutic Metallopharmaceuti-cals. Chem. Rev. 1999, 99, 2511–2533. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, U.K.; Gupta, M.; Karki, S.S.; Battacharya, S.; Rathinasamy, S.; Sivakumar, T. Synthesis and pharmacological activities of some mononuclear Ru(II) complexes. Bioorg. Med. Chem. 2005, 13, 5766–5773. [Google Scholar] [CrossRef] [PubMed]
- Schluga, P.; Hartinger, C.; Egger, A.; Reisner, E.; Galanski, M.; Jakupec, M.A.; Keppler, B.K. Redox behavior of tumor-inhibiting ruthenium (III) complexes and effects of physiological reductants on their binding to GMP. Dalton Trans. 2006, 14, 1796–1802. [Google Scholar] [CrossRef] [PubMed]
- Önfelt, B.; Lincoln, P.; Nordén, B. Femtosecond linear dichroism of DNA-intercalating chromophores: Solvation and charge separation dynamics of [Ru(phen)2dppz]2+ systems. Proc. Natl. Acad. Sci. USA 2000, 97, 5708–5713. [Google Scholar] [CrossRef] [PubMed]
- Önfelt, B.; Lincoln, P.; Nordén, B. Enantioselective DNA threading dynamics by phenazine-linked [Ru(phen)2dppz]2+ dimers. J. Am. Chem. Soc. 2001, 123, 3630–3637. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, C.; Haq, I.; Thomas, J.A. A Facile Route to Bimetallic Ruthenium Dipyrido-phenazine Complexes. Inorg. Chem. 2004, 43, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Pierard, F.; Mesmaeker, A.K.-D. Bifunctional transition metal as nucleic acid photoprobes and photoreagents. Inorg. Chem. Commun. 2006, 9, 111–126. [Google Scholar] [CrossRef]
- Wilhelmsson, L.M.; Westerlund, F.; Lincoln, P.; Nordén, B. DNA-Binding of Semi-Rigid Binuclear Ruthenium Complex Δ,Δ-[µ-11-11′-bidppz)(phen)4Ru2]4+: Extremely Slow Intercalation Kinetics. J. Am. Chem. Soc. 2002, 124, 12092–12093. [Google Scholar] [CrossRef] [PubMed]
- Nordell, P.; Westerlund, F.; Wilhelmsson, L.M.; Nordén, B.; Lincoln, P. Kinetic Recognition of AT-Rich DNA by Ruthenium Complexes. Angew. Chem. Int. Ed. 2007, 46, 2203–2206. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.J.; Kwon, B.H.; Choi, B.H.; Bae, C.H.; Seo, M.S.; Nam, W.; Kim, S.K. Intercalation of bulky Δ,Δ- and Λ,Λ-bis-Ru(II) complex between DNA base pairs. J. Inorg. Biochem. 2008, 102, 1885–1891. [Google Scholar] [CrossRef] [PubMed]
- Kwon, B.H.; Choi, B.H.; Lee, H.M.; Jang, Y.J.; Lee, J.C.; Kim, S.K. Binding Modes of New Bis-Ru(II) Complexes to DNA: Effect of the Length of the Linker. Bull. Korean Chem. Soc. 2010, 31, 1615–1620. [Google Scholar] [CrossRef]
- Duval-Valentine, G.; Thuong, N.T.; Hélène, C. Specific inhibition of transcription by triple helix-forming. Proc. Natl. Acad. Sci. USA 1992, 89, 504–508. [Google Scholar] [CrossRef]
- Giovannangeli, C.; Thuong, N.T.; Hélène, C. Specific inhibition of transcription by triple helix-forming oligonucleotides. Proc. Natl. Acad. Sci. USA 1993, 90, 10013–10017. [Google Scholar] [CrossRef] [PubMed]
- Hanvey, J.C.; Shimizu, M.; Wells, R.D. Site-specific inhibition of EcoRI restriction/modifi-cation enzymes by a DNA triple helix. Nucleic Acid Res. 1990, 18, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Johnston, B.H. The S1-sensitive form of d(C-T)n·d(A-G)n: Chemical evidence for a three-stranded structure in plasmids. Science 1988, 241, 1791–1796. [Google Scholar] [CrossRef]
- Fulton, A.B. How crowded is the cytoplasm? Cell 1982, 30, 345–347. [Google Scholar] [CrossRef]
- Moon, J.H.; Kim, S.K.; Sehlstedt, U.; Rodger, A.; Nordén, B. DNA structural features responsible for sequence-dependent binding geometries of Hoechst 33258. Biopolymers 1996, 38, 593–606. [Google Scholar] [CrossRef]
- Tuite, E.; Nordén, B. Intercalative interactions of ethidium dyes with triplex structures. Bioorg. Med. Chem. 1995, 3, 701–711. [Google Scholar] [CrossRef]
- Youn, M.R.; Moon, S.J.; Lee, B.W.; Lee, D.J.; Kim, J.M.; Kim, S.K.; Lee, C.S. DNA Mediated Energy Transfer from 4′,6-Diamidino-2-phenylindole to Ru (II)[(1,10-phenanthroline)2L]2+: Effect of Ligand Structure. Bull. Korean Chem. Soc. 2005, 26, 537–542. [Google Scholar]
- Lincoln, P.; Broo, A.; Norden, B. Diastereomeric DNA-Binding Geometries of Intercalated Ruthenium(II) Trischelates Probed by Linear Dichroism: [Ru(phen)2DPPZ]2+ and [Ru(phen)2BDPPZ]2+. J. Chem. Soc. 1996, 118, 2644–2653. [Google Scholar] [CrossRef]
- Lee, B.W.; Moon, S.J.; Youn, M.R.; Kim, J.H.; Jang, H.G.; Kim, S.K. DNA Mediated Resonance Energy Transfer from 4′,6-Diamidino-2-Phenylindole to [Ru(1,10-Phenanthroline)2L]2+. Biophys. J. 2003, 85, 3865–3871. [Google Scholar] [CrossRef]
- Yun, B.H.; Kim, J.O.; Lee, B.W.; Lincoln, P.; Nordén, B.; Kim, J.M.; Kim, S.K. Simultaneous binding of Ru(II)[(1,10-phenanthroline)2dipyridophenazine]2+ and minor groove binder 4′,6-diamino-2-phenylindole to poly[d(A-T)2] at high binding densities. J. Phys. Chem. B 2003, 107, 9858–9864. [Google Scholar] [CrossRef]
- Nair, R.B.; Cullum, B.M.; Murphy, C.J. Optical Properties of [Ru(phen)2dppz]2+ as a Function of Nonaqueous Environment. Inorg. Chem. 1997, 36, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, Y.; Friedman, A.E.; Turro, N.J.; Barton, J.K. Characterization of dipyridophenazine complexes of ruthenium(II)—The light switch effect as a function of nucleic-acid sequence and conformation. Biochemistry 1992, 31, 10809–10816. [Google Scholar] [CrossRef] [PubMed]
- Maheswari, P.U.; Rajendiran, V.; Palaniandavar, M.; Parthasarathi, R.; Subramanian, V.J. Synthesis, characterization and DNA-binding properties of rac-[Ru(5,6-dmp)2(dppz)]2+-enantiopreferential DNA binding and co-ligand promoted exciton coupling. J. Inorg. Biochem. 2006, 100, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Plenum Press: New York, NY, USA, 1990; p. 248. [Google Scholar]
- Hiort, C.; Nordén, B.; Rodger, A. Enantiopreferential DNA binding of [RuII(phenanthroline)3]2+ studied with linear and circular dichroism. J. Am.Chem. Soc. 1990, 112, 1971–1982. [Google Scholar] [CrossRef]







© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barai, H.R.; Lee, D.J.; Han, S.W.; Jang, Y.J. Interaction and Binding Modes of bis-Ruthenium(II) Complex to Synthetic DNAs. Metals 2016, 6, 141. https://doi.org/10.3390/met6060141
Barai HR, Lee DJ, Han SW, Jang YJ. Interaction and Binding Modes of bis-Ruthenium(II) Complex to Synthetic DNAs. Metals. 2016; 6(6):141. https://doi.org/10.3390/met6060141
Chicago/Turabian StyleBarai, Hasi Rani, Dong Jin Lee, Sung Wook Han, and Yoon Jung Jang. 2016. "Interaction and Binding Modes of bis-Ruthenium(II) Complex to Synthetic DNAs" Metals 6, no. 6: 141. https://doi.org/10.3390/met6060141