Influenza-Omics and the Host Response: Recent Advances and Future Prospects
Abstract
:1. Introduction
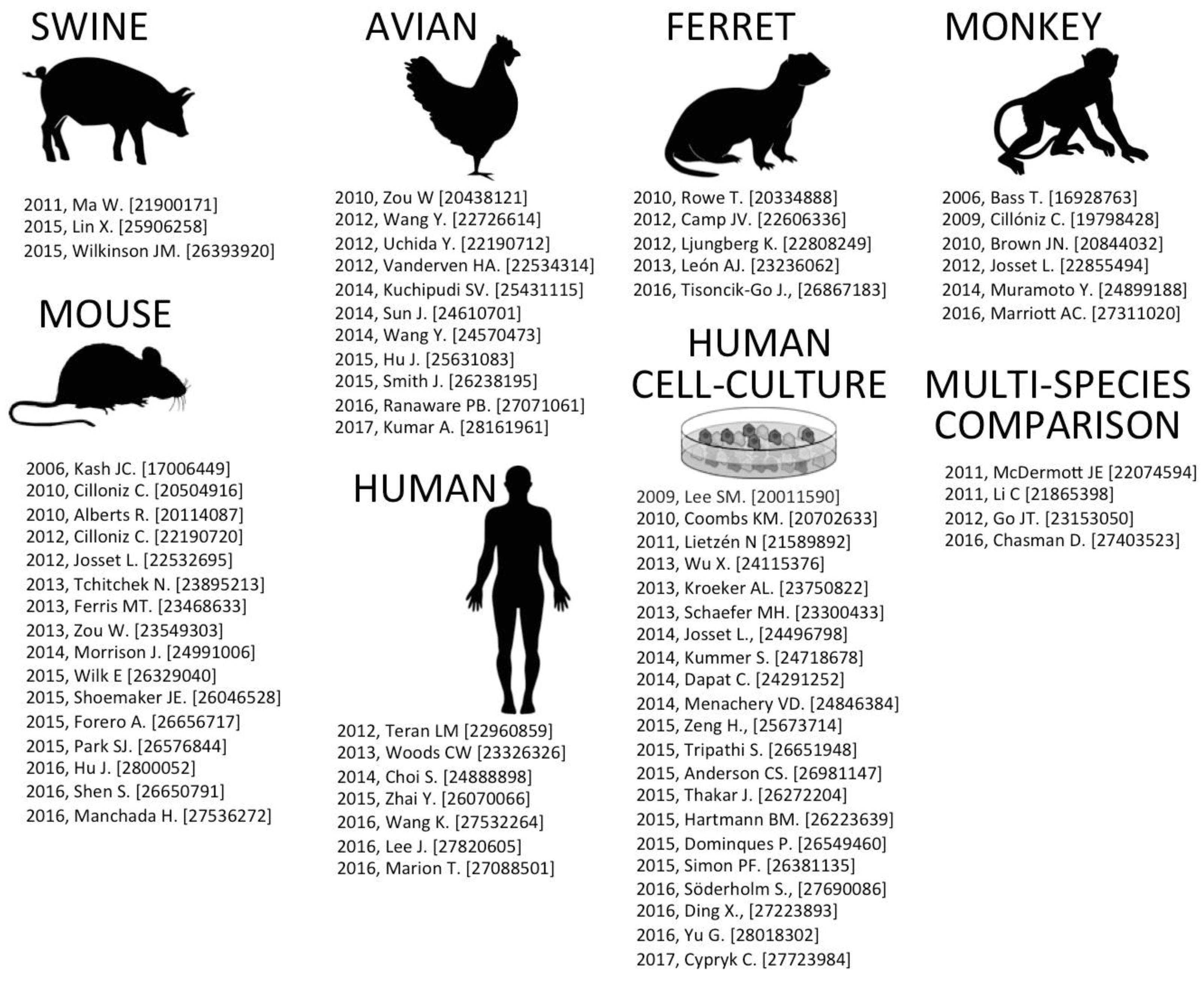
2. Host Response Models of IAV Infection
2.1. Swine-IAV Infection
2.2. Avian-IAV Infection
2.3. Ferret-IAV Infection
2.4. Monkey (Macaque)-IAV Infection
2.5. Mouse-IAV Infection
2.6. Human-IAV Infection
2.7. Human Cell Culture-IAV Infection
3. Discussion
Acknowledgments
Conflicts of Interest
References
- Oxford, K.L.; Wendler, J.P.; McDermott, J.E.; White, R.A., III; Powell, J.D.; Jacobs, J.M.; Adkins, J.N.; Waters, K.M. The landscape of viral proteomics and its potential to impact human health. Expert Rev. Proteom. 2016, 13, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Aevermann, B.D.; Pickett, B.E.; Kumar, S.; Klem, E.B.; Agnihothram, S.; Askovich, P.S.; Bankhead, A., 3rd; Bolles, M.; Carter, V.; Chang, J.; et al. A comprehensive collection of systems biology data characterizing the host response to viral infection. Sci. Data 2014, 1, 140033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Aevermann, B.D.; Anderson, T.K.; Burke, D.F.; Dauphin, G.; Gu, Z.; He, S.; Kumar, S.; Larsen, C.N.; Lee, A.J.; et al. Influenza research database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. 2017, 45, D466–D474. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Edgar, R. Ncbi geo: Mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 2007, 35, D760–D765. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. Geoquery: A bridge between the gene expression omnibus (geo) and bioconductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Keenliside, J. Pandemic influenza a H1N1 in swine and other animals. Curr. Top. Microbiol. Immunol. 2013, 370, 259–271. [Google Scholar] [PubMed]
- Vincent, A.; Awada, L.; Brown, I.; Chen, H.; Claes, F.; Dauphin, G.; Donis, R.; Culhane, M.; Hamilton, K.; Lewis, N.; et al. Review of influenza a virus in swine worldwide: A call for increased surveillance and research. Zoonoses Public Health 2014, 61, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Schultz-Cherry, S.; Olsen, C.W.; Easterday, B.C. History of swine influenza. Curr. Top. Microbiol. Immunol. 2013, 370, 21–28. [Google Scholar] [PubMed]
- Ma, W.; Belisle, S.E.; Mosier, D.; Li, X.; Stigger-Rosser, E.; Liu, Q.; Qiao, C.; Elder, J.; Webby, R.; Katze, M.G.; et al. 2009 pandemic H1N1 influenza virus causes disease and upregulation of genes related to inflammatory and immune responses, cell death, and lipid metabolism in pigs. J. Virol. 2011, 85, 11626–11637. [Google Scholar] [CrossRef] [PubMed]
- Go, J.T.; Belisle, S.E.; Tchitchek, N.; Tumpey, T.M.; Ma, W.; Richt, J.A.; Safronetz, D.; Feldmann, H.; Katze, M.G. 2009 pandemic H1N1 influenza virus elicits similar clinical course but differential host transcriptional response in mouse, macaque, and swine infection models. BMC Genom. 2012, 13, 627. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Huang, C.; Shi, J.; Wang, R.; Sun, X.; Liu, X.; Zhao, L.; Jin, M. Investigation of pathogenesis of H1N1 influenza virus and swine Streptococcus suis serotype 2 co-infection in pigs by microarray analysis. PLoS ONE 2015, 10, e0124086. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, H.; Wen, Z.; Wu, S.; Huang, C.; Jia, G.; Chen, H.; Jin, M. Transcription analysis on response of swine lung to H1N1 swine influenza virus. BMC Genom. 2011, 12, 398. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.M.; Gunvaldsen, R.E.; Detmer, S.E.; Dyck, M.K.; Dixon, W.T.; Foxcroft, G.R.; Plastow, G.S.; Harding, J.C. Transcriptomic and epigenetic profiling of the lung of influenza-infected pigs: A comparison of different birth weight and susceptibility groups. PLoS ONE 2015, 10, e0138653. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zou, W.; Jia, G.; Zhou, H.; Hu, Y.; Peng, M.; Chen, H.; Jin, M. Analysis of cellular proteome alterations in porcine alveolar macrophage cells infected with 2009 (H1N1) and classical swine H1N1 influenza viruses. J. Proteom. 2012, 75, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Bi, Y.; Wong, G.; Gray, G.C.; Gao, G.F.; Li, S. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J. Virol. 2015, 89, 8671–8676. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Brahmakshatriya, V.; Lupiani, B.; Reddy, S.M.; Soibam, B.; Benham, A.L.; Gunaratne, P.; Liu, H.C.; Trakooljul, N.; Ing, N.; et al. Integrated analysis of microrna expression and mrna transcriptome in lungs of avian influenza virus infected broilers. BMC Genom. 2012, 13, 278. [Google Scholar] [CrossRef] [PubMed]
- Ranaware, P.B.; Mishra, A.; Vijayakumar, P.; Gandhale, P.N.; Kumar, H.; Kulkarni, D.D.; Raut, A.A. Genome wide host gene expression analysis in chicken lungs infected with avian influenza viruses. PLoS ONE 2016, 11, e0153671. [Google Scholar] [CrossRef] [PubMed]
- Uchida, Y.; Watanabe, C.; Takemae, N.; Hayashi, T.; Oka, T.; Ito, T.; Saito, T. Identification of host genes linked with the survivability of chickens infected with recombinant viruses possessing H5N1 surface antigens from a highly pathogenic avian influenza virus. J. Virol. 2012, 86, 2686–2695. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Smith, N.; Yu, L.; Paton, I.R.; Gutowska, M.W.; Forrest, H.L.; Danner, A.F.; Seiler, J.P.; Digard, P.; Webster, R.G.; et al. A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genom. 2015, 16, 574. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Han, Z.; Shao, Y.; Cao, Z.; Kong, X.; Liu, S. Comparative proteome analysis of tracheal tissues in response to infectious bronchitis coronavirus, newcastle disease virus, and avian influenza virus H9 subtype virus infection. Proteomics 2014, 14, 1403–1423. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Ke, J.; Zhang, A.; Zhou, M.; Liao, Y.; Zhu, J.; Zhou, H.; Tu, J.; Chen, H.; Jin, M. Proteomics analysis of differential expression of chicken brain tissue proteins in response to the neurovirulent H5N1 avian influenza virus infection. J. Proteome Res. 2010, 9, 3789–3798. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.S.; Bohm, R.; Hartley-Tassell, L.E.; Steen, J.A.; Wang, H.; Lukowski, S.W.; Hawthorne, P.L.; Trezise, A.E.; Coloe, P.J.; Grimmond, S.M.; et al. Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza a virus receptors. Nat. Commun. 2014, 5, 5750. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, N.M. Animal models for influenza virus transmission studies: A historical perspective. Curr. Opin. Virol. 2015, 13, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Tisoncik-Go, J.; Gasper, D.J.; Kyle, J.E.; Eisfeld, A.J.; Selinger, C.; Hatta, M.; Morrison, J.; Korth, M.J.; Zink, E.M.; Kim, Y.M.; et al. Integrated omics analysis of pathogenic host responses during pandemic H1N1 influenza virus infection: The crucial role of lipid metabolism. Cell Host Microbe 2016, 19, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Leon, A.J.; Banner, D.; Xu, L.; Ran, L.; Peng, Z.; Yi, K.; Chen, C.; Xu, F.; Huang, J.; Zhao, Z.; et al. Sequencing, annotation, and characterization of the influenza ferret infectome. J. Virol. 2013, 87, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, K.; McBrayer, A.; Camp, J.V.; Chu, Y.K.; Tapp, R.; Noah, D.L.; Grimes, S.; Proctor, M.L.; Liljestrom, P.; Jonsson, C.B.; et al. Host gene expression signatures discriminate between ferrets infected with genetically similar H1N1 strains. PLoS ONE 2012, 7, e40743. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.V.; Svensson, T.L.; McBrayer, A.; Jonsson, C.B.; Liljestrom, P.; Bruder, C.E. De-novo transcriptome sequencing of a normalized cdna pool from influenza infected ferrets. PLoS ONE 2012, 7, e37104. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.S.; Taubenberger, J.K.; Bray, M. The use of nonhuman primates in research on seasonal, pandemic and avian influenza, 1893–2014. Antivir. Res. 2015, 117, 75–98. [Google Scholar] [CrossRef] [PubMed]
- Baas, T.; Baskin, C.R.; Diamond, D.L.; Garcia-Sastre, A.; Bielefeldt-Ohmann, H.; Tumpey, T.M.; Thomas, M.J.; Carter, V.S.; Teal, T.H.; Van Hoeven, N.; et al. Integrated molecular signature of disease: Analysis of influenza virus-infected macaques through functional genomics and proteomics. J. Virol. 2006, 80, 10813–10828. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.N.; Palermo, R.E.; Baskin, C.R.; Gritsenko, M.; Sabourin, P.J.; Long, J.P.; Sabourin, C.L.; Bielefeldt-Ohmann, H.; Garcia-Sastre, A.; Albrecht, R.; et al. Macaque proteome response to highly pathogenic avian influenza and 1918 reassortant influenza virus infections. J. Virol. 2010, 84, 12058–12068. [Google Scholar] [CrossRef] [PubMed]
- Cilloniz, C.; Shinya, K.; Peng, X.; Korth, M.J.; Proll, S.C.; Aicher, L.D.; Carter, V.S.; Chang, J.H.; Kobasa, D.; Feldmann, F.; et al. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 2009, 5, e1000604. [Google Scholar] [CrossRef] [PubMed]
- Josset, L.; Engelmann, F.; Haberthur, K.; Kelly, S.; Park, B.; Kawoaka, Y.; Garcia-Sastre, A.; Katze, M.G.; Messaoudi, I. Increased viral loads and exacerbated innate host responses in aged macaques infected with the 2009 pandemic H1N1 influenza a virus. J. Virol. 2012, 86, 11115–11127. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.E.; Shankaran, H.; Eisfeld, A.J.; Belisle, S.E.; Neuman, G.; Li, C.; McWeeney, S.; Sabourin, C.; Kawaoka, Y.; Katze, M.G.; et al. Conserved host response to highly pathogenic avian influenza virus infection in human cell culture, mouse and macaque model systems. BMC Syst. Biol. 2011, 5, 190. [Google Scholar] [CrossRef] [PubMed]
- Marriott, A.C.; Dennis, M.; Kane, J.A.; Gooch, K.E.; Hatch, G.; Sharpe, S.; Prevosto, C.; Leeming, G.; Zekeng, E.G.; Staples, K.J.; et al. Influenza a virus challenge models in cynomolgus macaques using the authentic inhaled aerosol and intra-nasal routes of infection. PLoS ONE 2016, 11, e0157887. [Google Scholar] [CrossRef] [PubMed]
- Forero, A.; Tisoncik-Go, J.; Watanabe, T.; Zhong, G.; Hatta, M.; Tchitchek, N.; Selinger, C.; Chang, J.; Barker, K.; Morrison, J.; et al. The 1918 influenza virus PB2 protein enhances virulence through the disruption of inflammatory and wnt-mediated signaling in mice. J. Virol. 2015, 90, 2240–2253. [Google Scholar] [CrossRef] [PubMed]
- Cilloniz, C.; Pantin-Jackwood, M.J.; Ni, C.; Goodman, A.G.; Peng, X.; Proll, S.C.; Carter, V.S.; Rosenzweig, E.R.; Szretter, K.J.; Katz, J.M.; et al. Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection. J. Virol. 2010, 84, 7613–7624. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Chen, D.; Xiong, M.; Zhu, J.; Lin, X.; Wang, L.; Zhang, J.; Chen, L.; Zhang, H.; Chen, H.; et al. Insights into the increasing virulence of the swine-origin pandemic H1N1/2009 influenza virus. Sci. Rep. 2013, 3, 1601. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kumar, M.; Kwon, H.I.; Seong, R.K.; Han, K.; Song, J.M.; Kim, C.J.; Choi, Y.K.; Shin, O.S. Dynamic changes in host gene expression associated with H5N8 avian influenza virus infection in mice. Sci. Rep. 2015, 5, 16512. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.; Josset, L.; Tchitchek, N.; Chang, J.; Belser, J.A.; Swayne, D.E.; Pantin-Jackwood, M.J.; Tumpey, T.M.; Katze, M.G. H7N9 and other pathogenic avian influenza viruses elicit a three-pronged transcriptomic signature that is reminiscent of 1918 influenza virus and is associated with lethal outcome in mice. J. Virol. 2014, 88, 10556–10568. [Google Scholar] [CrossRef] [PubMed]
- Alberts, R.; Srivastava, B.; Wu, H.; Viegas, N.; Geffers, R.; Klawonn, F.; Novoselova, N.; do Valle, T.Z.; Panthier, J.J.; Schughart, K. Gene expression changes in the host response between resistant and susceptible inbred mouse strains after influenza a infection. Microbes Infect. 2010, 12, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Wilk, E.; Pandey, A.K.; Leist, S.R.; Hatesuer, B.; Preusse, M.; Pommerenke, C.; Wang, J.; Schughart, K. Rnaseq expression analysis of resistant and susceptible mice after influenza a virus infection identifies novel genes associated with virus replication and important for host resistance to infection. BMC Genom. 2015, 16, 655. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.P.; Welsh, C.E. Informatics resources for the collaborative cross and related mouse populations. Mamm. Genome 2015, 26, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Ferris, M.T.; Aylor, D.L.; Bottomly, D.; Whitmore, A.C.; Aicher, L.D.; Bell, T.A.; Bradel-Tretheway, B.; Bryan, J.T.; Buus, R.J.; Gralinski, L.E.; et al. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2013, 9, e1003196. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Gao, Z.; Wang, X.; Gu, M.; Liang, Y.; Liu, X.; Hu, S.; Liu, H.; Liu, W.; Chen, S.; et al. Itraq-based quantitative proteomics reveals important host factors involved in the high pathogenicity of the h5n1 avian influenza virus in mice. Med. Microbiol. Immunol. 2016, 206, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Li, J.; Hilchey, S.; Shen, X.; Tu, C.; Qiu, X.; Ng, A.; Ghaemmaghami, S.; Wu, H.; Zand, M.S.; et al. Ion-current-based temporal proteomic profiling of influenza-a-virus-infected mouse lungs revealed underlying mechanisms of altered integrity of the lung microvascular barrier. J. Proteome Res. 2016, 15, 540–553. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Liang, L.; Li, Y.; Liu, L.; Guan, Y.; Jiang, Y.; Chen, H. Proteomic analysis of the lungs of mice infected with different pathotypes of H5N1 avian influenza viruses. Proteomics 2012, 12, 1970–1982. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.W.; McClain, M.T.; Chen, M.; Zaas, A.K.; Nicholson, B.P.; Varkey, J.; Veldman, T.; Kingsmore, S.F.; Huang, Y.; Lambkin-Williams, R.; et al. A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza h1n1 or H3N2. PLoS ONE 2013, 8, e52198. [Google Scholar] [CrossRef] [PubMed]
- Teran, L.M.; Ruggeberg, S.; Santiago, J.; Fuentes-Arenas, F.; Hernandez, J.L.; Montes-Vizuet, A.R.; Xinping, L.; Franz, T. Immune response to seasonal influenza a virus infection: A proteomic approach. Arch. Med. Res. 2012, 43, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Langevin, S.; O'Hern, C.S.; Shattuck, M.D.; Ogle, S.; Forero, A.; Morrison, J.; Slayden, R.; Katze, M.G.; Kirby, M. Anomaly detection in host signaling pathways for the early prognosis of acute infection. PLoS ONE 2016, 11, e0160919. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Franco, L.M.; Atmar, R.L.; Quarles, J.M.; Arden, N.; Bucasas, K.L.; Wells, J.M.; Nino, D.; Wang, X.; Zapata, G.E.; et al. Host transcriptional response to influenza and other acute respiratory viral infections—A prospective cohort study. PLoS Pathog. 2015, 11, e1004869. [Google Scholar] [CrossRef] [PubMed]
- Marion, T.; Elbahesh, H.; Thomas, P.G.; DeVincenzo, J.P.; Webby, R.; Schughart, K. Respiratory mucosal proteome quantification in human influenza infections. PLoS ONE 2016, 11, e0153674. [Google Scholar] [CrossRef] [PubMed]
- Josset, L.; Zeng, H.; Kelly, S.M.; Tumpey, T.M.; Katze, M.G. Transcriptomic characterization of the novel avian-origin influenza a (H7N9) virus: Specific host response and responses intermediate between avian (H5N1 and H7N7) and human (H3N2) viruses and implications for treatment options. MBio 2014, 5, e01102–e01113. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Belser, J.A.; Goldsmith, C.S.; Gustin, K.M.; Veguilla, V.; Katz, J.M.; Tumpey, T.M. A(H7N9) virus results in early induction of proinflammatory cytokine responses in both human lung epithelial and endothelial cells and shows increased human adaptation compared with avian H5N1 virus. J. Virol. 2015, 89, 4655–4667. [Google Scholar] [CrossRef] [PubMed]
- Coombs, K.M.; Berard, A.; Xu, W.; Krokhin, O.; Meng, X.; Cortens, J.P.; Kobasa, D.; Wilkins, J.; Brown, E.G. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J. Virol. 2010, 84, 10888–10906. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Gardy, J.L.; Cheung, C.Y.; Cheung, T.K.; Hui, K.P.; Ip, N.Y.; Guan, Y.; Hancock, R.E.; Peiris, J.S. Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages. PLoS ONE 2009, 4, e8072. [Google Scholar] [CrossRef] [PubMed]
- Cypryk, W.; Lorey, M.; Puustinen, A.; Nyman, T.A.; Matikainen, S. Proteomic and bioinformatic characterization of extracellular vesicles released from human macrophages upon influenza a virus infection. J. Proteome Res. 2017, 16, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Lietzen, N.; Ohman, T.; Rintahaka, J.; Julkunen, I.; Aittokallio, T.; Matikainen, S.; Nyman, T.A. Quantitative subcellular proteome and secretome profiling of influenza a virus-infected human primary macrophages. PLoS Pathog. 2011, 7, e1001340. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Green, J.; Pollard, J.J.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Korth, M.J.; Tchitchek, N.; Benecke, A.G.; Katze, M.G. Systems approaches to influenza-virus host interactions and the pathogenesis of highly virulent and pandemic viruses. Semin. Immunol. 2013, 25, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Kollmus, H.; Wilk, E.; Schughart, K. Systems biology and systems genetics-novel innovative approaches to study host-pathogen interactions during influenza infection. Curr. Opin. Virol. 2014, 6, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. MMBR 2012, 76, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Tchitchek, N.; Eisfeld, A.J.; Tisoncik-Go, J.; Josset, L.; Gralinski, L.E.; Becavin, C.; Tilton, S.C.; Webb-Robertson, B.J.; Ferris, M.T.; Totura, A.L.; et al. Specific mutations in H5N1 mainly impact the magnitude and velocity of the host response in mice. BMC Syst. Biol. 2013, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.T.; Tchitchek, N.; Ghosh, D.; Benecke, A.; Katze, M.G. A chemokine gene expression signature derived from meta-analysis predicts the pathogenicity of viral respiratory infections. BMC Syst. Biol. 2011, 5, 202. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Chan, E.Y.; Li, Y.; Diamond, D.L.; Korth, M.J.; Katze, M.G. Virus-host interactions: From systems biology to translational research. Curr. Opin. Microbiol. 2009, 12, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Garcia-Sastre, A. Antiviral innate immunity through the lens of systems biology. Virus Res. 2016, 218, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Munk, C.; Sommer, A.F.; Konig, R. Systems-biology approaches to discover anti-viral effectors of the human innate immune response. Viruses 2011, 3, 1112–1130. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Powell, J.D.; Waters, K.M. Influenza-Omics and the Host Response: Recent Advances and Future Prospects. Pathogens 2017, 6, 25. https://doi.org/10.3390/pathogens6020025
Powell JD, Waters KM. Influenza-Omics and the Host Response: Recent Advances and Future Prospects. Pathogens. 2017; 6(2):25. https://doi.org/10.3390/pathogens6020025
Chicago/Turabian StylePowell, Joshua D., and Katrina M. Waters. 2017. "Influenza-Omics and the Host Response: Recent Advances and Future Prospects" Pathogens 6, no. 2: 25. https://doi.org/10.3390/pathogens6020025




