Soluble CD14 as a Diagnostic Biomarker for Smear-Negative HIV-Associated Tuberculosis
Abstract
:1. Introduction
2. Results
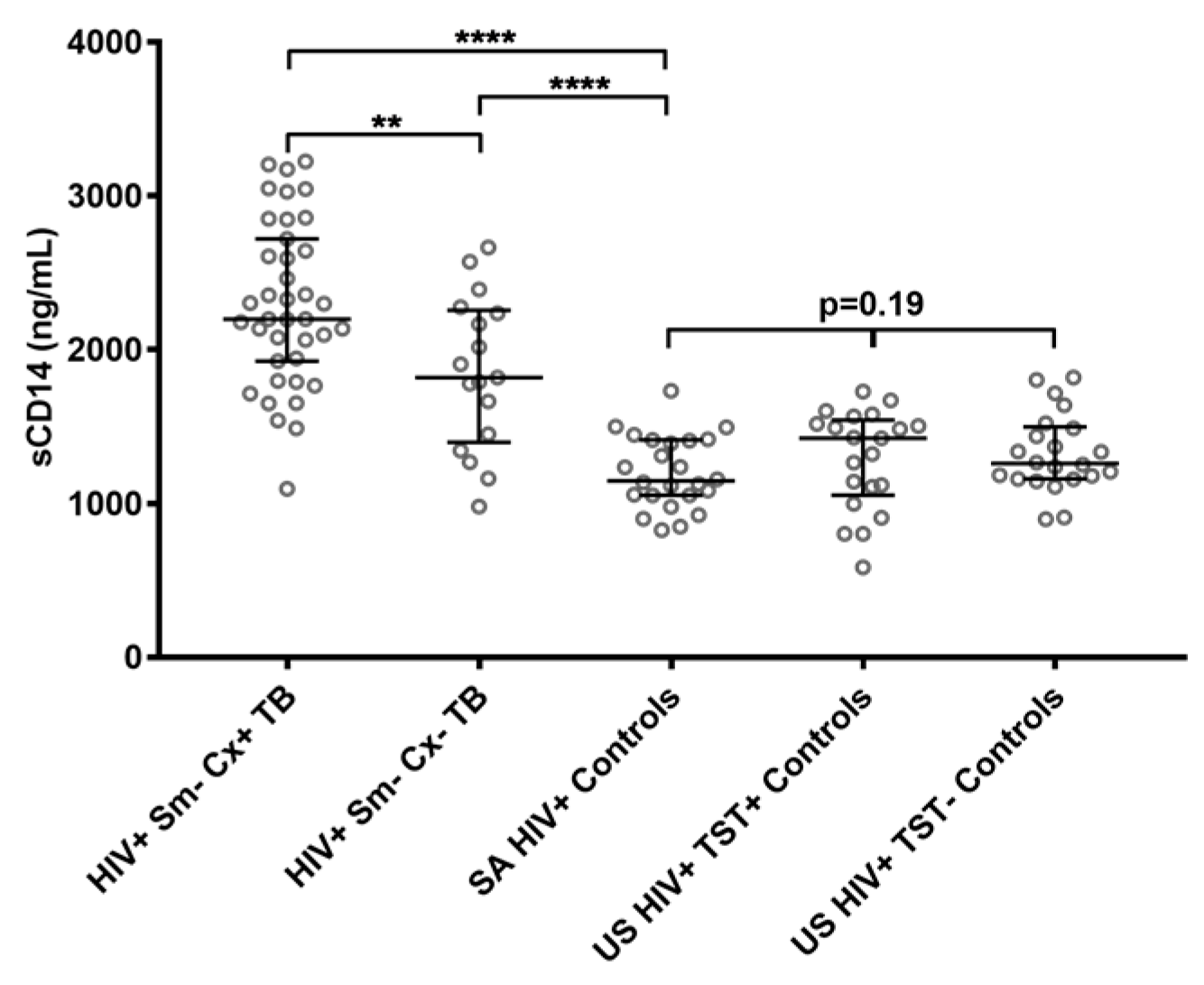
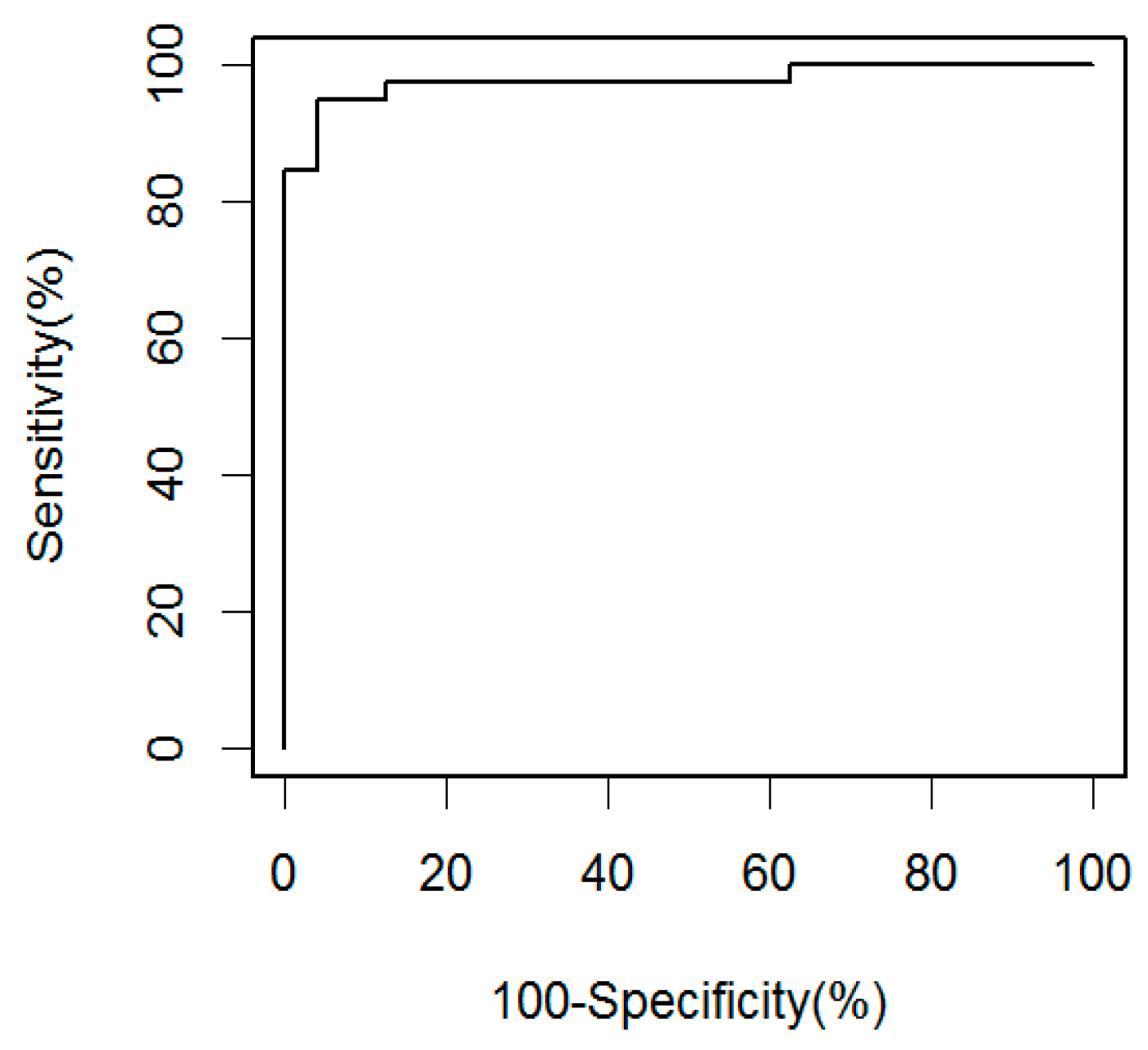
2.1. Soluble CD14 Serum Concentrations with Sensitivity and Specificity for Smear-Negative HIV-Associated TB
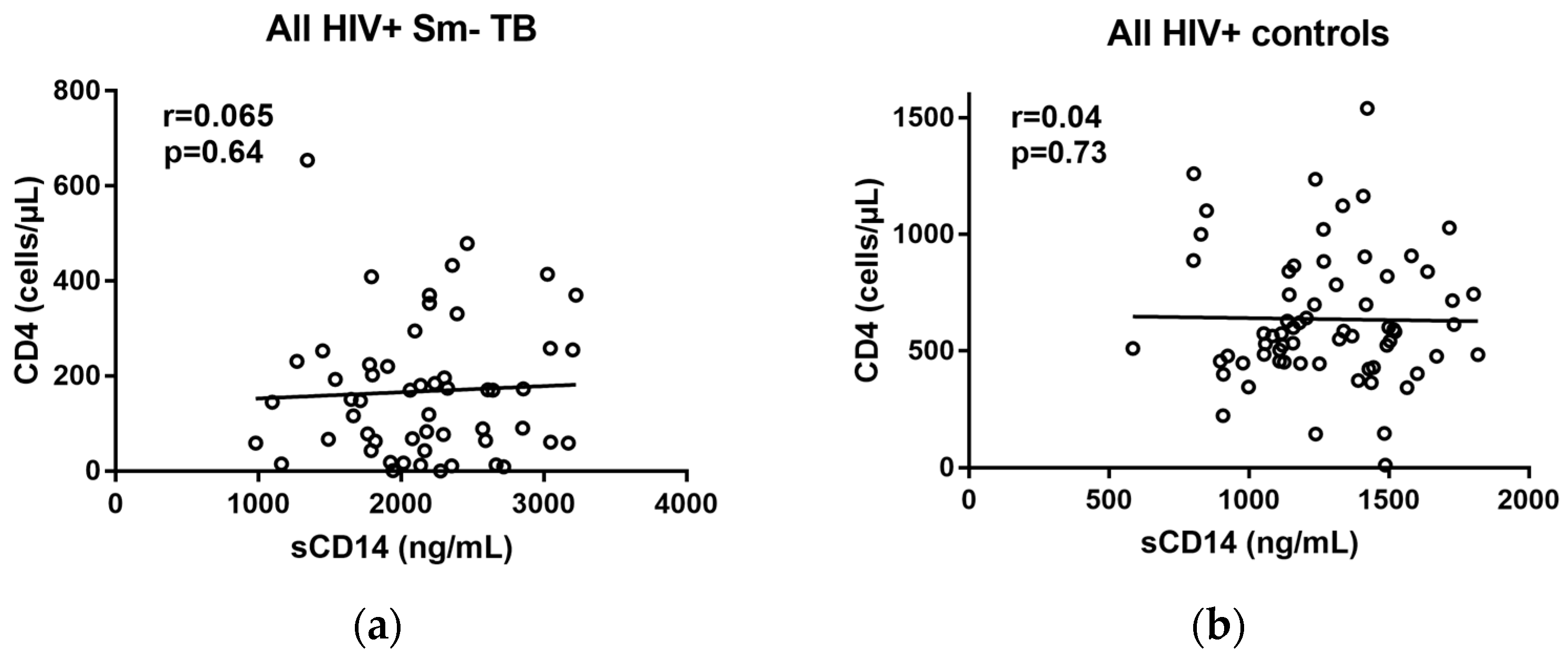
2.2. Correlations of sCD14 and CD4 Levels
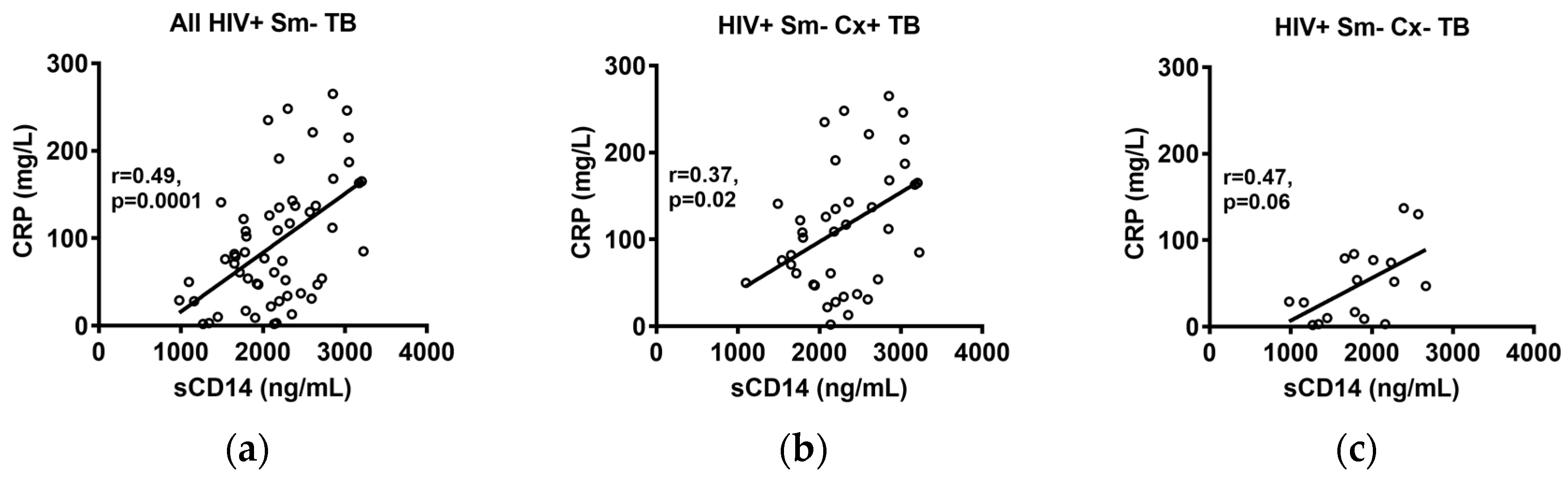
2.3. Correlations of sCD14 and CRP Levels
3. Discussion
4. Materials and Methods
4.1. Study Design and Subjects
4.2. Sample Preparation
4.3. ELISAs
4.4. CRP Measurements
4.5. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Barry, C.E., 3rd; Boshoff, H.I.; Dartois, V.; Dick, T.; Ehrt, S.; Flynn, J.; Schnappinger, D.; Wilkinson, R.J.; Young, D. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 2009, 7, 845–855. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guidelines on the Management of Latent Tuberculosis Infection; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Williams, B.G.; Dye, C. Antiretroviral drugs for tuberculosis control in the era of HIV/AIDS. Science 2003, 301, 1535–1537. [Google Scholar] [CrossRef] [PubMed]
- Akolo, C.; Adetifa, I.; Shepperd, S.; Volmink, J. Treatment of latent tuberculosis infection in HIV infected persons. Cochrane Database Syst. Rev. 2010. [Google Scholar] [CrossRef] [PubMed]
- Achkar, J.M.; Jenny-Avital, E.R. Incipient and subclinical tuberculosis: Defining early disease states in the context of host immune response. J. Infect. Dis. 2011, 204 (Suppl. 4), S1179–S1186. [Google Scholar] [CrossRef] [PubMed]
- Achkar, J.M.; Lawn, S.D.; Moosa, M.Y.; Wright, C.A.; Kasprowicz, V.O. Adjunctive tests for diagnosis of tuberculosis: Serology, ELISPOT for site-specific lymphocytes, urinary lipoarabinomannan, string test, and fine needle aspiration. J. Infect. Dis. 2011, 204 (Suppl. 4), S1130–S1141. [Google Scholar] [CrossRef] [PubMed]
- Golden, M.P.; Vikram, H.R. Extrapulmonary tuberculosis: An overview. Am. Fam. Physician 2005, 72, 1761–1768. [Google Scholar] [PubMed]
- Bhattacharya, D.; Danaviah, S.; Muema, D.M.; Akilimali, N.A.; Moodley, P.; Ndung’u, T.; Das, G. Cellular architecture of spinal granulomas and the immunological response in Tuberculosis Patients coinfected with HIV. Front. Immunol. 2017, 8, 1120. [Google Scholar] [CrossRef] [PubMed]
- Reid, M.J.; Shah, N.S. Approaches to tuberculosis screening and diagnosis in people with HIV in resource-limited settings. Lancet Infect. Dis. 2009, 9, 173–184. [Google Scholar] [CrossRef]
- Bassett, I.V.; Wang, B.; Chetty, S.; Giddy, J.; Losina, E.; Mazibuko, M.; Bearnot, B.; Allen, J.; Walensky, R.P.; Freedberg, K.A. Intensive tuberculosis screening for HIV-infected patients starting antiretroviral therapy in Durban, South Africa. Clin. Infect. Dis. 2010, 51, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Pai, N.P.; Vadnais, C.; Denkinger, C.; Engel, N.; Pai, M. Point-of-care testing for infectious diseases: Diversity, complexity, and barriers in low-and middle-income countries. PLoS Med. 2012, 9, e1001306. [Google Scholar] [CrossRef] [PubMed]
- Heidebrecht, C.L.; Podewils, L.J.; Pym, A.S.; Cohen, T.; Mthiyane, T.; Wilson, D. Assessing the utility of Xpert® MTB/RIF as a screening tool for patients admitted to medical wards in South Africa. Sci. Rep. 2016, 6, 19391. [Google Scholar] [CrossRef] [PubMed]
- Denkinger, C.M.; Kik, S.V.; Cirillo, D.M.; Casenghi, M.; Shinnick, T.; Weyer, K.; Gilpin, C.; Boehme, C.C.; Schito, M.; Kimerling, M. Defining the needs for next generation assays for tuberculosis. J. Infect. Dis. 2015, 211 (Suppl. 2), S29–S38. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Hanrahan, C.; Wang, Z.Y.; Dendukuri, N.; Lawn, S.D.; Denkinger, C.M.; Steingart, K.R. Lateral Flow Urine Lipoarabinomannan Assay for Detecting Active Tuberculosis in HIV-Positive Adults. Cochrane Database Syst. Rev. 2016. [Google Scholar] [CrossRef]
- WHO. The Use of Lateral Flow Urine Lipoarabinomannan Assay (LF-LAM) for the Diagnosis and Screening of Active Tuberculosis in People Living with HIV: Policy Guidance; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Agassandian, M.; Shurin, G.V.; Ma, Y.; Shurin, M.R. C-reactive protein and lung diseases. Int. J. Biochem. Cell Biol. 2014, 53, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Abernethy, T.J.; Avery, O.T. The occurrence during acute infections of a protein not normally present in the blood. J. Exp. Med. 1941, 73, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Hurlimann, J.; Thorbecke, G.; Hochwald, G. The liver as the site of C-reactive protein formation. J. Exp. Med. 1966, 123, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Black, S.; Kushner, I.; Samols, D. C-reactive Protein. J. Biol. Chem. 2004, 279, 48487–48490. [Google Scholar] [CrossRef] [PubMed]
- Windgassen, E.B.; Funtowicz, L.; Lunsford, T.N.; Harris, L.A.; Mulvagh, S.L. C-reactive protein and high-sensitivity C-reactive protein: An update for clinicians. Postgrad. Med. 2011, 123, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Badri, M.; Maartens, G. Performance of serum C-reactive protein as a screening test for smear-negative tuberculosis in an ambulatory high HIV prevalence population. PLoS ONE 2011, 6, e15248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chegou, N.N.; Sutherland, J.S.; Malherbe, S.; Crampin, A.C.; Corstjens, P.L.; Geluk, A.; Mayanja-Kizza, H.; Loxton, A.G.; van der Spuy, G.; Stanley, K. Diagnostic performance of a seven-marker serum protein biosignature for the diagnosis of active TB disease in African primary healthcare clinic attendees with signs and symptoms suggestive of TB. Thorax 2016, 71, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Drain, P.K.; Mayeza, L.; Bartman, P.; Hurtado, R.; Moodley, P.; Varghese, S.; Maartens, G.; Alvarez, G.G.; Wilson, D. Diagnostic accuracy and clinical role of rapid C-reactive protein testing in HIV-infected individuals with presumed tuberculosis in South Africa. Int. J. Tuberc. Lung Dis. 2014, 18, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, R.; Malherbe, S.; Loxton, A.G.; Stanley, K.; van der Spuy, G.; Walzl, G.; Chegou, N.N. Identification of novel host biomarkers in plasma as candidates for the immunodiagnosis of tuberculosis disease and monitoring of tuberculosis treatment response. Oncotarget 2016, 7, 57581–57592. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Semitala, F.C.; Atuhumuza, E.; Katende, J.; Mwebe, S.; Asege, L.; Armstrong, D.T.; Andama, A.O.; Dowdy, D.W.; Davis, J.L. Point-of-care C-reactive protein-based tuberculosis screening for people living with HIV: A diagnostic accuracy study. Lancet Infect. Dis. 2017, 17, 1285–1292. [Google Scholar] [CrossRef]
- Lawn, S.D.; Kerkhoff, A.D.; Vogt, M.; Wood, R. Diagnostic and prognostic value of serum C-reactive protein for screening for HIV-associated tuberculosis. Int. J. Tuberc. Lung Dis. 2013, 17, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Chaisson, L.H.; Patel, S.M.; Allen, I.E.; Drain, P.K.; Wilson, D.; Cattamanchi, A. Diagnostic accuracy of C-reactive protein for active pulmonary tuberculosis: A meta-analysis. Int. J. Tuberc. Lung Dis. 2017, 21, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Frey, E.; Miller, D.; Jahr, T.G.; Sundan, A.; Bazil, V.; Espevik, T.; Finlay, B.B.; Wright, S. Soluble CD14 participates in the response of cells to lipopolysaccharide. J. Exp. Med. 1992, 176, 1665–1671. [Google Scholar] [CrossRef]
- Zhang, Y.; Doerfler, M.; Lee, T.C.; Guillemin, B.; Rom, W.N. Mechanisms of stimulation of interleukin-1 beta and tumor necrosis factor-alpha by Mycobacterium tuberculosis components. J. Clin. Investig. 1993, 91, 2076. [Google Scholar] [CrossRef] [PubMed]
- Pugin, J.; Heumann, D.; Tomasz, A.; Kravchenko, V.V.; Akamatsu, Y.; Nishijima, M.; Glauser, M.P.; Tobias, P.S.; Ulevitch, R.J. CD14 is a pattern recognition receptor. Immunity 1994, 1, 509–516. [Google Scholar] [CrossRef]
- Kol, A.; Lichtman, A.H.; Finberg, R.W.; Libby, P.; Kurt-Jones, E.A. Cutting edge: Heat shock protein (HSP) 60 activates the innate immune response: CD14 is an essential receptor for HSP60 activation of mononuclear cells. J. Immunol. 2000, 164, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Goyert, S.; Ferrero, E.; Seremetis, S.; Winchester, R.; Silver, J.; Mattison, A. Biochemistry and expression of myelomonocytic antigens. J. Immunol. 1986, 137, 3909–3914. [Google Scholar] [PubMed]
- Goyert, S.M.; Ferrero, E.; Rettig, W.J.; Yenamandra, A.K.; Obata, F.; Le Beau, M.M. The CD14 monocyte differentiation antigen maps to a region encoding growth factors and receptors. Science 1988, 239, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Bazil, V.; Strominger, J. Shedding as a mechanism of down-modulation of CD14 on stimulated human monocytes. J. Immunol. 1991, 147, 1567–1574. [Google Scholar] [PubMed]
- Labeta, M.O.; Durieux, J.J.; Fernandez, N.; Herrmann, R.; Ferrara, P. Release from a human monocyte-like cell line of two different soluble forms of the lipopolysaccharide receptor, CD14. Eur. J. Immunol. 1993, 23, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Dentener, M.; Bazil, V.; Von Asmuth, E.; Ceska, M.; Buurman, W. Involvement of CD14 in lipopolysaccharide-induced tumor necrosis factor-alpha, IL-6 and IL-8 release by human monocytes and alveolar macrophages. J. Immunol. 1993, 150, 2885–2891. [Google Scholar] [PubMed]
- Wright, S.D.; Ramos, R.A.; Tobias, P.S.; Ulevitch, R.J.; Mathison, J.C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 1990, 249, 1431–1433. [Google Scholar] [CrossRef] [PubMed]
- Lawn, S.D.; Labeta, M.O.; Arias, M.; Acheampong, J.W.; Griffin, G.E. Elevated serum concentrations of soluble CD14 in HIV(−) and HIV(+) patients with tuberculosis in Africa: Prolonged elevation during anti-tuberculosis treatment. Clin. Exp. Immunol. 2000, 120, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, Z.A.; Wong, E.B.; Ndung’u, T.; Kasprowicz, V.O.; Bishai, W.R. Latent and Active Tuberculosis Infection Increase Immune Activation in Individuals Co-Infected with HIV. EBioMedicine 2015, 2, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Wyndham-Thomas, C.; Corbière, V.; Selis, E.; Payen, M.-C.; Goffard, J.-C.; Van Vooren, J.-P.; Mascart, F.; Dirix, V. Immune Activation by Mycobacteriumtuberculosis in HIV-Infected and -Uninfected Subjects. J. Acquir. Immune Defici. Syndr. 2017, 74, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Feruglio, S.L.; Trøseid, M.; Damås, J.K.; Kvale, D.; Dyrhol-Riise, A.M. Soluble Markers of the Toll-Like Receptor 4 Pathway Differentiate between Active and Latent Tuberculosis and Are Associated with Treatment Responses. PLoS ONE 2013, 8, e69896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achkar, J.M.; Cortes, L.; Croteau, P.; Yanofsky, C.; Mentinova, M.; Rajotte, I.; Schirm, M.; Zhou, Y.; Junqueira-Kipnis, A.P.; Kasprowicz, V.O. Host protein biomarkers identify active tuberculosis in HIV uninfected and co-infected individuals. EBioMedicine 2015, 2, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Siev, M.; Wilson, D.; Kainth, S.; Kasprowicz, V.O.; Feintuch, C.M.; Jenny-Avital, E.R.; Achkar, J.M. Antibodies against Mycobacterial proteins as biomarkers for HIV-associated smear-negative tuberculosis. Clin. Vaccine Immunol. 2014, 21, 791–798. [Google Scholar] [CrossRef] [PubMed]
- WHO. Improving the Diagnosis and Treatment of Smear-Negative Pulmonary and Extrapulmonary Tuberculosis among Adults and Adolescents: Recommendations for HIV-Prevalent and Resource-Constrained Settings; World Health Organization: Geneva, Switzerland, 2007. [Google Scholar]
- Getahun, H.; Harrington, M.; O’Brien, R.; Nunn, P. Diagnosis of smear-negative pulmonary tuberculosis in people with HIV infection or AIDS in resource-constrained settings: Informing urgent policy changes. Lancet 2007, 369, 2042–2049. [Google Scholar] [CrossRef]
- Hargreaves, N.J.; Kadzakumanja, O.; Whitty, C.J.; Salaniponi, F.M.; Harries, A.D.; Squire, S.B. ‘Smear-negative’ pulmonary tuberculosis in a DOTS programme: Poor outcomes in an area of high HIV seroprevalence. Int. J. Tuberc. Lung Dis. 2001, 5, 847–854. [Google Scholar] [PubMed]
- Kang’ombe, C.T.; Harries, A.D.; Ito, K.; Clark, T.; Nyirenda, T.E.; Aldis, W.; Nunn, P.P.; Semba, R.D.; Salaniponi, F.M. Long-term outcome in patients registered with tuberculosis in Zomba, Malawi: Mortality at 7 years according to initial HIV status and type of TB. Int. J. Tuberc. Lung Dis. 2004, 8, 829–836. [Google Scholar] [PubMed]
- Ahn, J.S.; Choi, S.; Jang, S.H.; Chang, H.J.; Kim, J.H.; Nahm, K.B.; Oh, S.W.; Choi, E.Y. Development of a point-of-care assay system for high-sensitivity C-reactive protein in whole blood. Clin. Chim. Acta 2003, 332, 51–59. [Google Scholar] [CrossRef]
- Fernández-Sánchez, C.; McNeil, C.J.; Rawson, K.; Nilsson, O.; Leung, H.Y.; Gnanapragasam, V. One-step immunostrip test for the simultaneous detection of free and total prostate specific antigen in serum. J. Immunol. Methods 2005, 307, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Yuhi, T.; Nagatani, N.; Endo, T.; Kerman, K.; Takamura, Y.; Tamiya, E. A novel enhancement assay for immunochromatographic test strips using gold nanoparticles. Anal. Bioanal. Chem. 2006, 385, 1414–1420. [Google Scholar] [CrossRef] [PubMed]
- Dempfle, C.-E.; Schraml, M.; Besenthal, I.; Hansen, R.; Gehrke, J.; Korte, W.; Risch, M.; Quehenberger, P.; Handler, S.; Minar, E.; et al. Multicentre evaluation of a new point-of-care test for the quantitative determination of D-dimer. Clin. Chim. Acta 2001, 307, 211–218. [Google Scholar] [CrossRef]
- McNerney, R.; Daley, P. Towards a point-of-care test for active tuberculosis: Obstacles and opportunities. Nat. Rev. Microbiol. 2011, 9, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Gubala, V.; Harris, L.F.; Ricco, A.J.; Tan, M.X.; Williams, D.E. Point of care diagnostics: Status and future. Anal. Chem. 2011, 84, 487–515. [Google Scholar] [CrossRef] [PubMed]
- Haziot, A.; Chen, S.; Ferrero, E.; Low, M.; Silber, R.; Goyert, S. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J. Immunol. 1988, 141, 547–552. [Google Scholar] [PubMed]
- Kedzierska, K.; Crowe, S.M. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr. Med. Chem. 2002, 9, 1893–1903. [Google Scholar] [CrossRef] [PubMed]
- Nockher, W.A.; Bergmann, L.; Scherberich, J.E. Increased soluble CD14 serum levels and altered CD14 expression of peripheral blood monocytes in HIV-infected patients. Clin. Exp. Immunol. 1994, 98, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Orenstein, J.M.; Fox, C.; Wahl, S.M. Macrophages as a source of HIV during opportunistic infections. Science 1997, 276, 1857–1861. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, J.M.; Douek, D.C. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 2008, 1, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, J.M.; Price, D.A.; Schacker, T.W.; Asher, T.E.; Silvestri, G.; Rao, S.; Kazzaz, Z.; Bornstein, E.; Lambotte, O.; Altmann, D. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006, 12, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Sandler, N.G.; Wand, H.; Roque, A.; Law, M.; Nason, M.C.; Nixon, D.E.; Pedersen, C.; Ruxrungtham, K.; Lewin, S.R.; Emery, S.; et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J. Infect. Dis. 2011, 203, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Shivakoti, R.; Gupta, A.; Ray, J.C.; Uprety, P.; Gupte, N.; Bhosale, R.; Mave, V.; Patil, S.; Balasubramanian, U.; Kinikar, A.; et al. Soluble CD14: An Independent Biomarker for the Risk of Mother-to-Child Transmission of HIV in a Setting of Preexposure and Postexposure Antiretroviral Prophylaxis. J. Infect. Dis. 2016, 213, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.; Aukrust, P.; Sundan, A.; Muller, F.; Froland, S.S.; Espevik, T. Elevated levels of serum-soluble CD14 in human immunodeficiency virus type 1 (HIV-1) infection: Correlation to disease progression and clinical events. Blood 1998, 92, 2084–2092. [Google Scholar] [PubMed]
- Thiebaut, R.; Charpentier, C.; Damond, F.; Taieb, A.; Antoine, R.; Capeau, J.; Chene, G.; Collin, G.; Matheron, S.; Descamps, D.; et al. Association of soluble CD14 and inflammatory biomarkers with HIV-2 disease progression. J. Infect. Dis. 2012, 55, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Duan, Z.; Li, D.; Li, D.; Wang, Z.; Ren, L.; Shen, T.; Shao, Y. Higher levels of circulating monocyte-platelet aggregates are correlated with viremia and increased sCD163 levels in HIV-1 infection. Cell. Mol. Immunol. 2015, 12, 435–543. [Google Scholar] [CrossRef] [PubMed]
- Krastinova, E.; Lecuroux, C.; Leroy, C.; Seng, R.; Cabie, A.; Rami, A.; Venet, A.; Meyer, L.; Goujard, C.; Cohort, A.P. High Soluble CD14 Levels at Primary HIV-1 Infection Predict More Rapid Disease Progression. J. Infect. Dis. 2015, 212, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Bastard, J.P.; Soulie, C.; Fellahi, S.; Haim-Boukobza, S.; Simon, A.; Katlama, C.; Calvez, V.; Marcelin, A.G.; Capeau, J. Circulating interleukin-6 levels correlate with residual HIV viraemia and markers of immune dysfunction in treatment-controlled HIV-infected patients. Antivir. Ther. 2012, 17, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Screening for tuberculosis and tuberculous infection in high-risk populations. Recommendations of the Advisory Committee for Elimination of Tuberculosis. MMWR Recomm. Rep. 1990, 39, 1–7. Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/00001642.htm (accessed on 20 February 2018).
- Roberts, P.L. Virus inactivation by solvent/detergent treatment using Triton X-100 in a high purity factor VIII. Biologicals 2008, 36, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
| Characteristic | Culture-Positive Subjects with TB (n = 39) | Culture-Negative Subjects with TB (n = 17) | p Value |
|---|---|---|---|
| Male sex (%) | 19 (49) | 8 (47) | 0.91 a |
| Age, mean years (±SD) | 33 (±7) | 32 (±6) | 0.86 b |
| CD4 cells/mm3, median (IQR) d | 119 (18–219) | 68 (15–193) | 0.41 c |
| CRP mg/L, median (IQR) | 109 (52–164) | 47 (10–77) | 0.001 c |
| History of prior TB (%) | 8 (21) | 1 (6) | 0.25 b |
| Characteristic | US HIV+ TST− (n = 22) | US HIV+ TST+ (n = 21) | SA HIV+ (n = 24) |
|---|---|---|---|
| Age, mean no. of years (±SD) | 48 (±10) | 46 (±13) | 33 (±7) |
| Male sex (%) | 10 (46) | 14 (64) | 6 (25) |
| CD4 cells/mm3, median (IQR) | 574 (449–744) | 525 (423–842) | 601 (483–742) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Ndumnego, O.C.; Chen, T.; Kim, R.S.; Jenny-Avital, E.R.; Ndung’u, T.; Wilson, D.; Achkar, J.M. Soluble CD14 as a Diagnostic Biomarker for Smear-Negative HIV-Associated Tuberculosis. Pathogens 2018, 7, 26. https://doi.org/10.3390/pathogens7010026
Liu Y, Ndumnego OC, Chen T, Kim RS, Jenny-Avital ER, Ndung’u T, Wilson D, Achkar JM. Soluble CD14 as a Diagnostic Biomarker for Smear-Negative HIV-Associated Tuberculosis. Pathogens. 2018; 7(1):26. https://doi.org/10.3390/pathogens7010026
Chicago/Turabian StyleLiu, Yanyan, Okechukwu C. Ndumnego, Tingting Chen, Ryung S. Kim, Elizabeth R. Jenny-Avital, Thumbi Ndung’u, Douglas Wilson, and Jacqueline M. Achkar. 2018. "Soluble CD14 as a Diagnostic Biomarker for Smear-Negative HIV-Associated Tuberculosis" Pathogens 7, no. 1: 26. https://doi.org/10.3390/pathogens7010026





