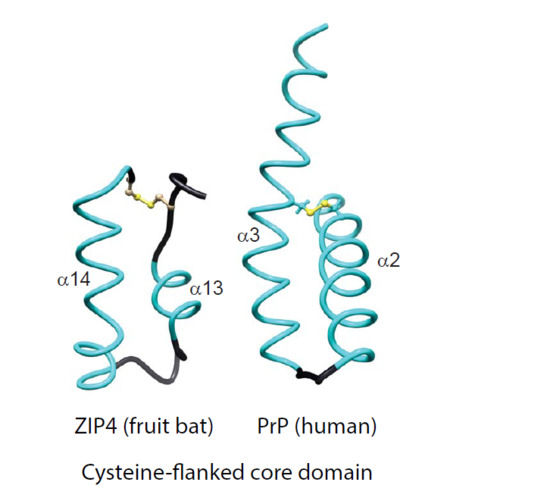
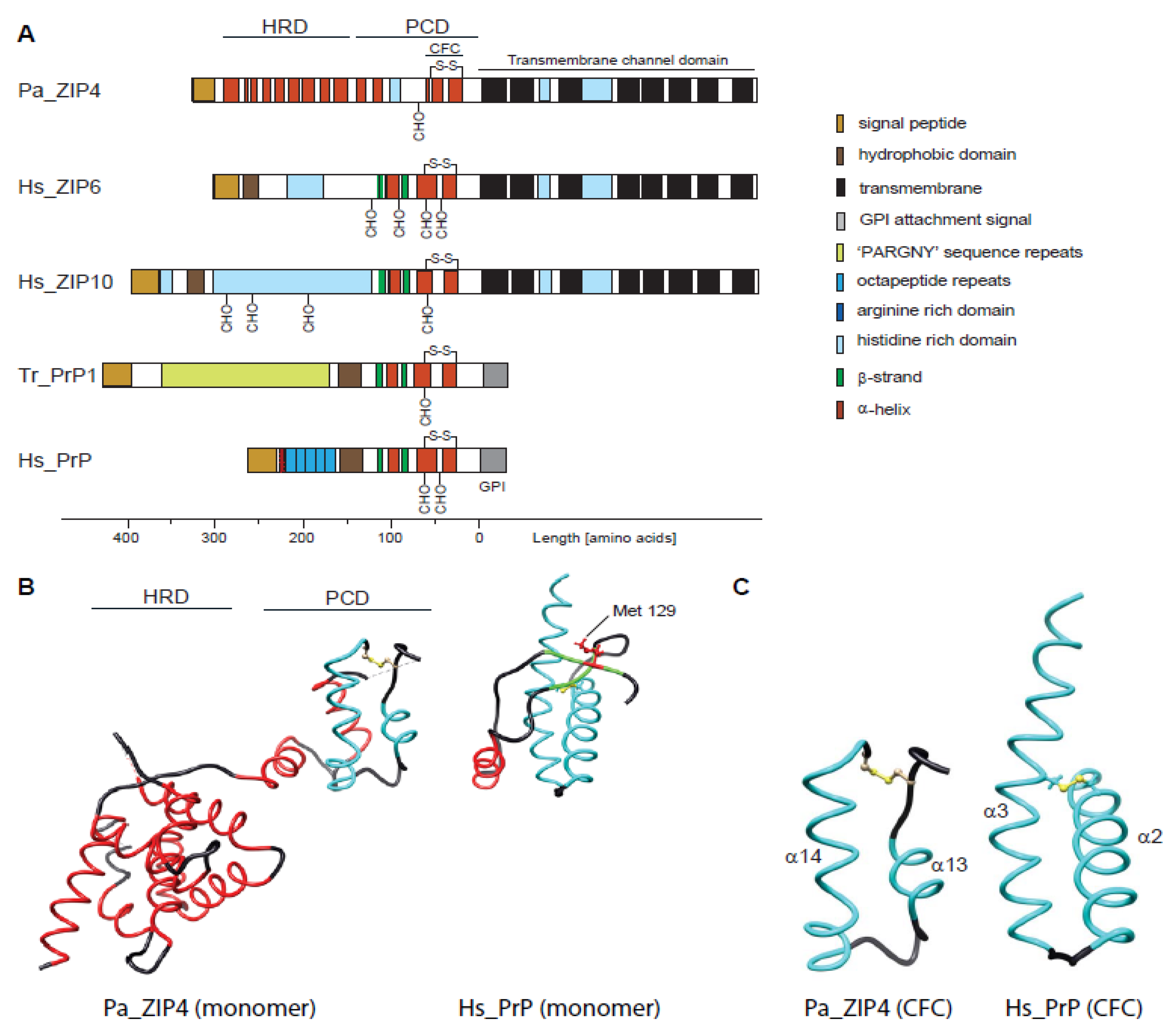
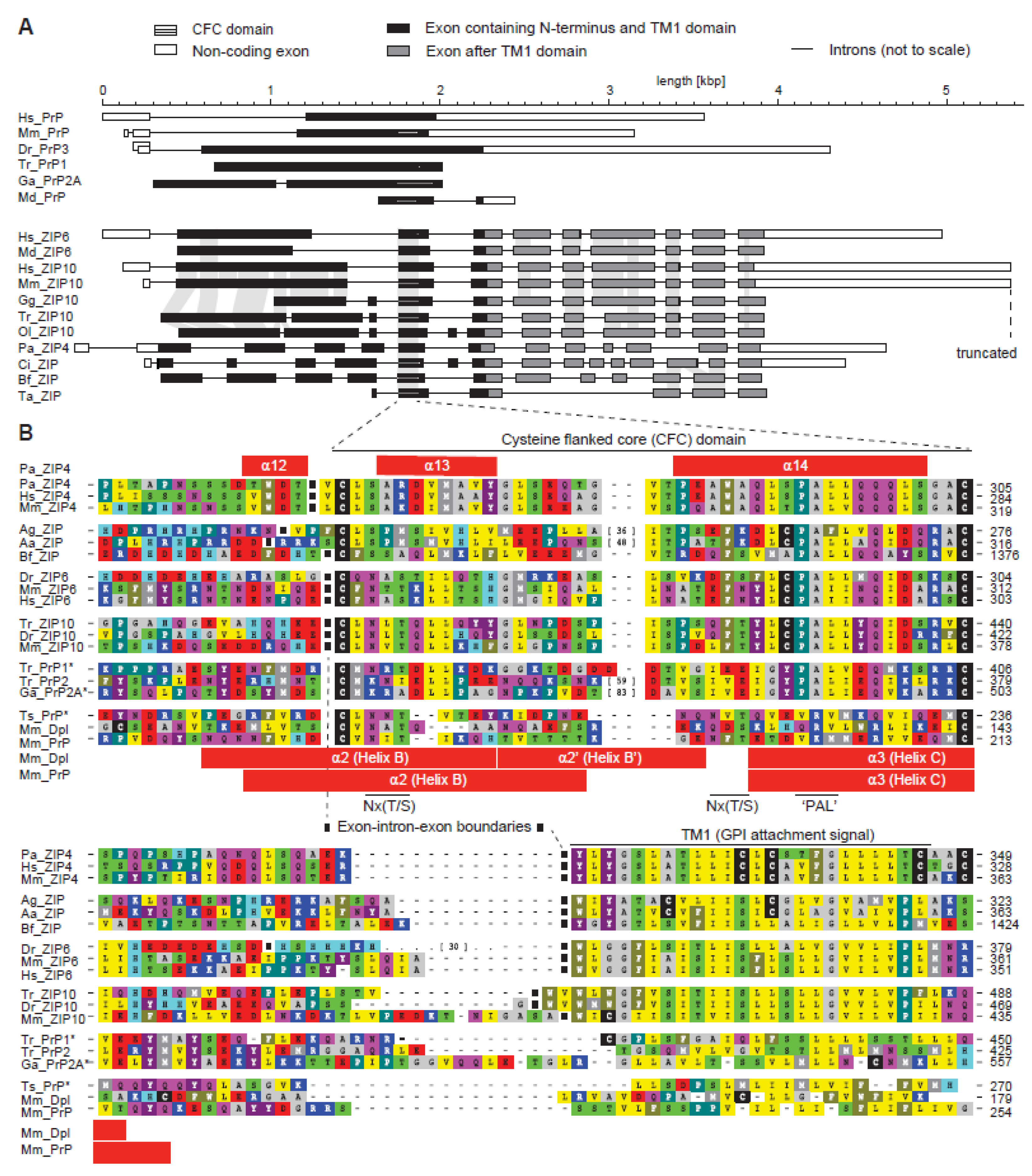
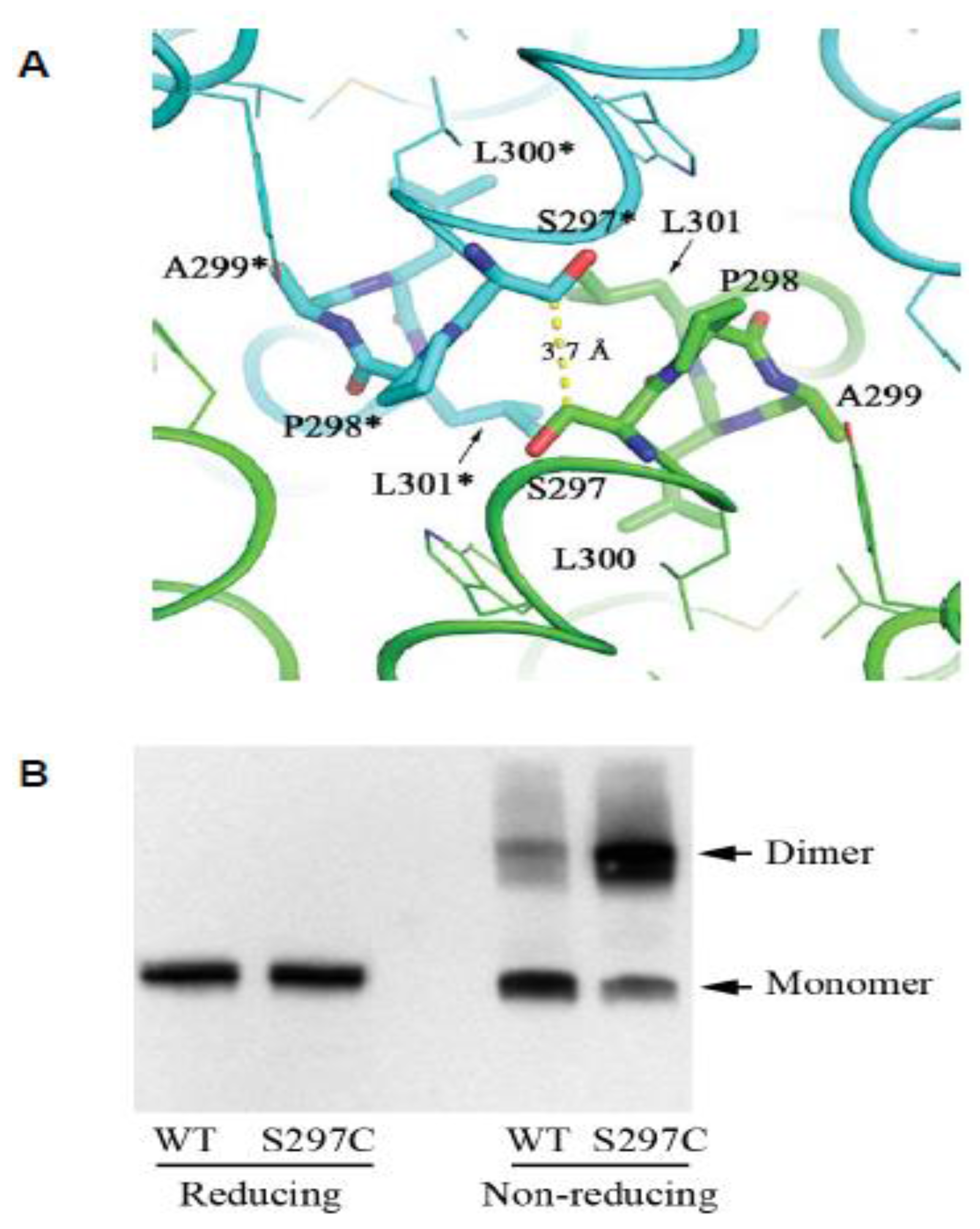
The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures
Abstract
:1. Introduction
2. Main
2.1. Comparison of Molecular Architectures of ZIP and Prion Proteins
2.2. Disappearance of CPAL Motif in the Prion Protein Subbranch of ZIP-PrP Protein Family
2.3. The CPAL Motif Represents a Dimerization Interface
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Aa | Anopheles aegypti |
| Ag | Anopheles gambiae |
| AE | Acrodermatitis enteropathica |
| Bf | Branchiostoma florida |
| CDF | cation diffusion facilitator |
| CFC | cysteine-flanked core |
| Ci | Ciona intestinalis |
| CTD | carboxy-terminal domain |
| Dm | Drosophila melanogaster |
| Dr | Danio rerio |
| ECD | extracellular domain |
| Ga | Gasterosteus aculateus |
| Gg | Gallus gallus |
| GPI | glycosylphosphatidylinositol |
| HRD | helix-rich domain |
| Hs | Homo sapiens |
| LZT | LIV-1 subfamily of ZIP zinc transporters |
| Md | Monodelphis domestica |
| Mm | Mus musculus |
| Ol | Oryzias latipes |
| ORF | open reading frame |
| Pa | Pteropus alecto |
| PCD | PAL-motif containing domain |
| PL | prion-like |
| PrPC | cellular prion protein |
| SLC | solute carrier |
| Ta | Trichoplax adhaerens |
| TM | transmembrane |
| Tr | Takifugu rubripes |
| Ts | Trachemys scripta |
References
- Prusiner, S.B. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B. Cell biology. A unifying role for prions in neurodegenerative diseases. Science 2012, 336, 1511–1513. [Google Scholar] [CrossRef] [PubMed]
- Sailer, A.; Bueler, H.; Fischer, M.; Aguzzi, A.; Weissmann, C. No propagation of prions in mice devoid of PrP. Cell 1994, 77, 967–968. [Google Scholar] [CrossRef]
- Moore, R.C.; Lee, I.Y.; Silverman, G.L.; Harrison, P.M.; Strome, R.; Heinrich, C.; Karunaratne, A.; Pasternak, S.H.; Chishti, M.A.; Liang, Y.; et al. Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J. Mol. Biol. 1999, 292, 797–817. [Google Scholar] [CrossRef] [PubMed]
- Silverman, G.L.; Qin, K.; Moore, R.C.; Yang, Y.; Mastrangelo, P.; Tremblay, P.; Prusiner, S.B.; Cohen, F.E.; Westaway, D. Doppel is an N-glycosylated, glycosylphosphatidylinositol-anchored protein. Expression in testis and ectopic production in the brains of Prnp(0/0) mice predisposed to Purkinje cell loss. J. Biol. Chem. 2000, 275, 26834–26841. [Google Scholar] [PubMed]
- Premzl, M.; Sangiorgio, L.; Strumbo, B.; Marshall Graves, J.A.; Simonic, T.; Gready, J.E. Shadoo, a new protein highly conserved from fish to mammals and with similarity to prion protein. Gene 2003, 314, 89–102. [Google Scholar] [CrossRef]
- Westaway, D.; Daude, N.; Wohlgemuth, S.; Harrison, P. The PrP-like proteins Shadoo and Doppel. Top. Curr. Chem. 2011, 305, 225–256. [Google Scholar] [PubMed]
- Watts, J.C.; Huo, H.; Bai, Y.; Ehsani, S.; Jeon, A.H.; Shi, T.; Daude, N.; Lau, A.; Young, R.; Xu, L.; et al. Interactome analyses identify ties of PrP and its mammalian paralogs to oligomannosidic N-glycans and endoplasmic reticulum-derived chaperones. PLoS Pathog. 2009, 5, e1000608. [Google Scholar] [CrossRef]
- Schmitt-Ulms, G.; Ehsani, S.; Watts, J.C.; Westaway, D.; Wille, H. Evolutionary descent of prion genes from the ZIP family of metal ion transporters. PLoS ONE 2009, 4, e7208. [Google Scholar] [CrossRef] [PubMed]
- Hatinen, T.; Holm, L.; Airaksinen, M.S. Loss of neurturin in frog—Comparative genomics study of GDNF family ligand-receptor pairs. Mol. Cell Neurosci. 2007, 34, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Airaksinen, M.S.; Holm, L.; Hatinen, T. Evolution of the GDNF family ligands and receptors. Brain Behav. Evol. 2006, 68, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.M.; Solomon, K.R.; Gold, J.P.; Tan, K.N. Cytoplasmic tail deletion of T cell receptor (TCR) beta-chain results in its surface expression as glycosylphosphatidylinositol-anchored polypeptide on mature T cells in the absence of TCR-alpha. J. Biol. Chem. 1994, 269, 22758–22763. [Google Scholar] [PubMed]
- Aguzzi, A.; Baumann, F.; Bremer, J. The Prion’s Elusive Reason for Being. Annu. Rev. Neurosci. 2008, 31, 439–477. [Google Scholar] [CrossRef] [PubMed]
- Castle, A.R.; Gill, A.C. Physiological Functions of the Cellular Prion Protein. Front. Mol. Biosci. 2017, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kim, B.E.; Dufner-Beattie, J.; Petris, M.J.; Andrews, G.; Eide, D.J. Acrodermatitis enteropathica mutations affect transport activity, localization and zinc-responsive trafficking of the mouse ZIP4 zinc transporter. Hum. Mol. Genet. 2004, 13, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Giunta, C.; Elcioglu, N.H.; Albrecht, B.; Eich, G.; Chambaz, C.; Janecke, A.R.; Yeowell, H.; Weis, M.; Eyre, D.R.; Kraenzlin, M.; et al. Spondylocheiro dysplastic form of the Ehlers-Danlos syndrome—An autosomal-recessive entity caused by mutations in the zinc transporter gene SLC39A13. Am. J. Hum. Genet. 2008, 82, 1290–1305. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.Y.; Duncan, F.E.; Que, E.L.; Kim, A.M.; O’Halloran, T.V.; Woodruff, T.K. Maternally-derived zinc transporters ZIP6 and ZIP10 drive the mammalian oocyte-to-egg transition. Mol. Hum. Reprod. 2014, 20, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.M.; Muraina, I.; Brethour, D.; Schmitt-Ulms, G.; Nimmanon, T.; Ziliotto, S.; Kille, P.; Hogstrand, C. Zinc transporter ZIP10 forms a heteromer with ZIP6 which regulates embryonic development and cell migration. Biochem. J. 2016, 473, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Miyagi, C.; Fukada, T.; Kagara, N.; Che, Y.S.; Hirano, T. Zinc transporter LIVI controls epithelial-mesenchymal transition in zebrafish gastrula organizer. Nature 2004, 429, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Malaga-Trillo, E.; Solis, G.P.; Schrock, Y.; Geiss, C.; Luncz, L.; Thomanetz, V.; Stuermer, C.A. Regulation of embryonic cell adhesion by the prion protein. PLoS Biol. 2009, 7, e55. [Google Scholar] [CrossRef] [PubMed]
- Brethour, D.; Mehrabian, M.; Williams, D.; Wang, X.; Ghodrati, F.; Ehsani, S.; Rubie, E.A.; Woodgett, J.R.; Sevalle, J.; Xi, Z.; et al. A ZIP6-ZIP10 heteromer controls NCAM1 phosphorylation and integration into focal adhesion complexes during epithelial-to-mesenchymal transition. Sci. Rep. 2017, 7, 40313. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Ulms, G.; Legname, G.; Baldwin, M.A.; Ball, H.L.; Bradon, N.; Bosque, P.J.; Crossin, K.L.; Edelman, G.M.; DeArmond, S.J.; Cohen, F.E.; et al. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J. Mol. Biol. 2001, 314, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- Slapsak, U.; Salzano, G.; Amin, L.; Abskharon, R.N.; Ilc, G.; Zupancic, B.; Biljan, I.; Plavec, J.; Giachin, G.; Legname, G. The N Terminus of the Prion Protein Mediates Functional Interactions with the Neuronal Cell Adhesion Molecule (NCAM) Fibronectin Domain. J. Biol. Chem. 2016, 291, 21857–21868. [Google Scholar] [CrossRef] [PubMed]
- Santuccione, A.; Sytnyk, V.; Leshchyns’ka, I.; Schachner, M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J. Cell Biol. 2005, 169, 341–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrabian, M.; Brethour, D.; Wang, H.; Xi, Z.; Rogaeva, E.; Schmitt-Ulms, G. The Prion Protein Controls Polysialylation of Neural Cell Adhesion Molecule 1 during Cellular Morphogenesis. PLoS ONE 2015, 10, e0133741. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Hornemann, S.; Wider, G.; Billeter, M.; Glockshuber, R.; Wuthrich, K. NMR structure of the mouse prion protein domain PrP(121-321). Nature 1996, 382, 180–182. [Google Scholar] [CrossRef] [PubMed]
- Wuthrich, K.; Riek, R. Three-dimensional structures of prion proteins. Adv. Protein Chem. 2001, 57, 55–82. [Google Scholar] [PubMed]
- Calzolai, L.; Lysek, D.A.; Perez, D.R.; Guntert, P.; Wuthrich, K. Prion protein NMR structures of chickens, turtles, and frogs. Proc. Natl. Acad. Sci. USA 2005, 102, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Christen, B.; Wuthrich, K.; Hornemann, S. Putative prion protein from Fugu (Takifugu rubripes). FEBS J. 2008, 275, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Sui, D.; Hu, J. Structural insights of ZIP4 extracellular domain critical for optimal zinc transport. Nat. Commun. 2016, 7, 11979. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, J.; Fellner, M.; Zhang, C.; Sui, D.; Hu, J. Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 2017, 3, e1700344. [Google Scholar] [CrossRef] [PubMed]
- Ehsani, S.; Tao, R.; Pocanschi, C.L.; Ren, H.; Harrison, P.M.; Schmitt-Ulms, G. Evidence for retrogene origins of the prion gene family. PLoS ONE 2011, 6, e26800. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.M.; Endo, T.; Colominas, C.; Groth, D.; Wheeler, S.F.; Harvey, D.J.; Wormald, M.R.; Serban, H.; Prusiner, S.B.; Kobata, A.; et al. Glycosylation differences between the normal and pathogenic prion protein isoforms. Proc. Natl. Acad. Sci. USA 1999, 96, 13044–13049. [Google Scholar] [CrossRef] [PubMed]
- Ehsani, S.; Salehzadeh, A.; Huo, H.; Reginold, W.; Pocanschi, C.L.; Ren, H.; Wang, H.; So, K.; Sato, C.; Mehrabian, M.; et al. LIV-1 ZIP ectodomain shedding in prion-infected mice resembles cellular response to transition metal starvation. J. Mol. Biol. 2012, 422, 556–574. [Google Scholar] [CrossRef] [PubMed]
- Pocanschi, C.L.; Ehsani, S.; Mehrabian, M.; Wille, H.; Reginold, W.; Trimble, W.S.; Wang, H.; Yee, A.; Arrowsmith, C.H.; Bozoky, Z.; et al. The ZIP5 Ectodomain Co-Localizes with PrP and May Acquire a PrP-Like Fold That Assembles into a Dimer. PLoS ONE 2013, 8, e72446. [Google Scholar] [CrossRef] [PubMed]
- Bin, B.H.; Fukada, T.; Hosaka, T.; Yamasaki, S.; Ohashi, W.; Hojyo, S.; Miyai, T.; Nishida, K.; Yokoyama, S.; Hirano, T. Biochemical characterization of human ZIP13 protein: A homo-dimerized zinc transporter involved in the Spondylocheiro dysplastic Ehlers-Danlos syndrome. J. Biol. Chem. 2011, 286, 40255–40265. [Google Scholar] [CrossRef] [PubMed]
- Priola, S.A.; Caughey, B.; Wehrly, K.; Chesebro, B. A 60-kDa prion protein (PrP) with properties of both the normal and scrapie-associated forms of PrP. J. Biol. Chem. 1995, 270, 3299–3305. [Google Scholar] [CrossRef] [PubMed]
- Knaus, K.J.; Morillas, M.; Swietnicki, W.; Malone, M.; Surewicz, W.K.; Yee, V.C. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 2001, 8, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Antony, L.; Hartmann, R.; Knaus, K.J.; Surewicz, K.; Surewicz, W.K.; Yee, V.C. Conformational diversity in prion protein variants influences intermolecular beta-sheet formation. EMBO J. 2010, 29, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.K.; Lustig, A.; Oesch, B.; Fatzer, R.; Zurbriggen, A.; Vandevelde, M. A monomer-dimer equilibrium of a cellular prion protein (PrPC) not observed with recombinant PrP. J. Biol. Chem. 2000, 275, 38081–38087. [Google Scholar] [CrossRef] [PubMed]
- Hundt, C.; Gauczynski, S.; Leucht, C.; Riley, M.L.; Weiss, S. Intra- and interspecies interactions between prion proteins and effects of mutations and polymorphisms. Biol. Chem. 2003, 384, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Beland, M.; Roucou, X. Homodimerization as a molecular switch between low and high efficiency PrP C cell surface delivery and neuroprotective activity. Prion 2013, 7, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Beland, M.; Motard, J.; Barbarin, A.; Roucou, X. PrP(C) homodimerization stimulates the production of PrPC cleaved fragments PrPN1 and PrPC1. J. Neurosci. 2012, 32, 13255–13263. [Google Scholar] [CrossRef] [PubMed]
- Supattapone, S.; Bosque, P.; Muramoto, T.; Wille, H.; Aagaard, C.; Peretz, D.; Nguyen, H.-O.B.; Heinrich, C.; Torchia, M.; Safar, J.; et al. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell 1999, 96, 869–878. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; Wille, H.; Schmitt-Ulms, G. The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures. Pathogens 2018, 7, 4. https://doi.org/10.3390/pathogens7010004
Hu J, Wille H, Schmitt-Ulms G. The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures. Pathogens. 2018; 7(1):4. https://doi.org/10.3390/pathogens7010004
Chicago/Turabian StyleHu, Jian, Holger Wille, and Gerold Schmitt-Ulms. 2018. "The Evolutionary unZIPping of a Dimerization Motif—A Comparison of ZIP and PrP Architectures" Pathogens 7, no. 1: 4. https://doi.org/10.3390/pathogens7010004