Biology of the Marine Heterotrophic Dinoflagellate Oxyrrhis marina: Current Status and Future Directions
Abstract
:1. Introduction
2. Taxonomy and Phylogeny of Oxyrrhis
3. Unusual Cytological and Genetic Features
3.1. Morphology
3.2. Nuclear, Mitochondrial and Plastid Genomes
4. Nutritional Modes
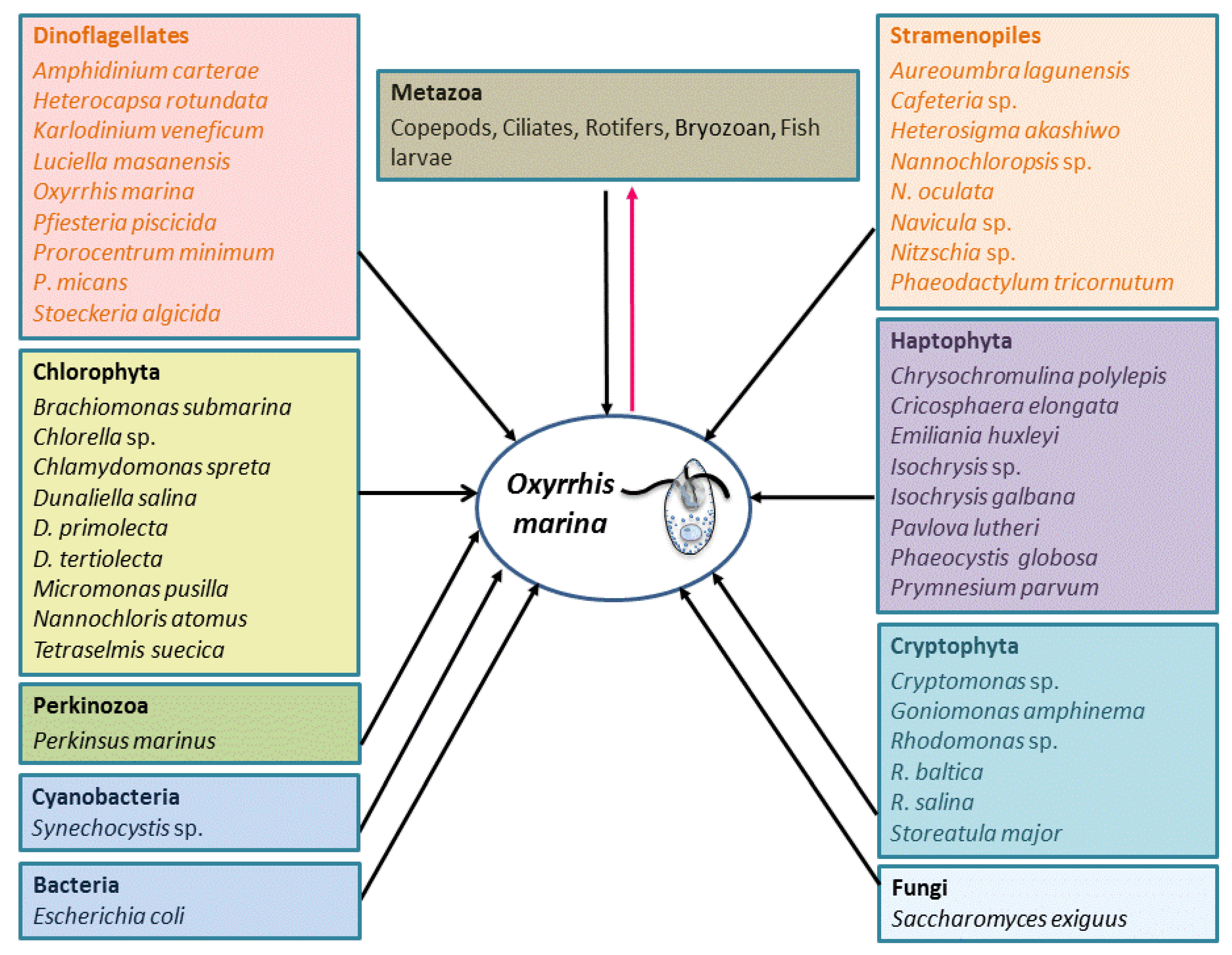
4.1. Phagotrophy
4.2. Saprotrophy
4.3. Cannibalization
4.4. Phototaxis, Circadian Rhythm, and the Potential of Solar Energy Utilization
5. Swimming and Feeding Behaviors
5.1. Swimming Behavior
5.2. Feeding Behavior
6. Life Cycle and Cell Cycle
6.1. Life Cycle
6.2. Cell Cycle
7. Physiology
8. Geographic Distribution
9. Future Directions for Research on O. marina
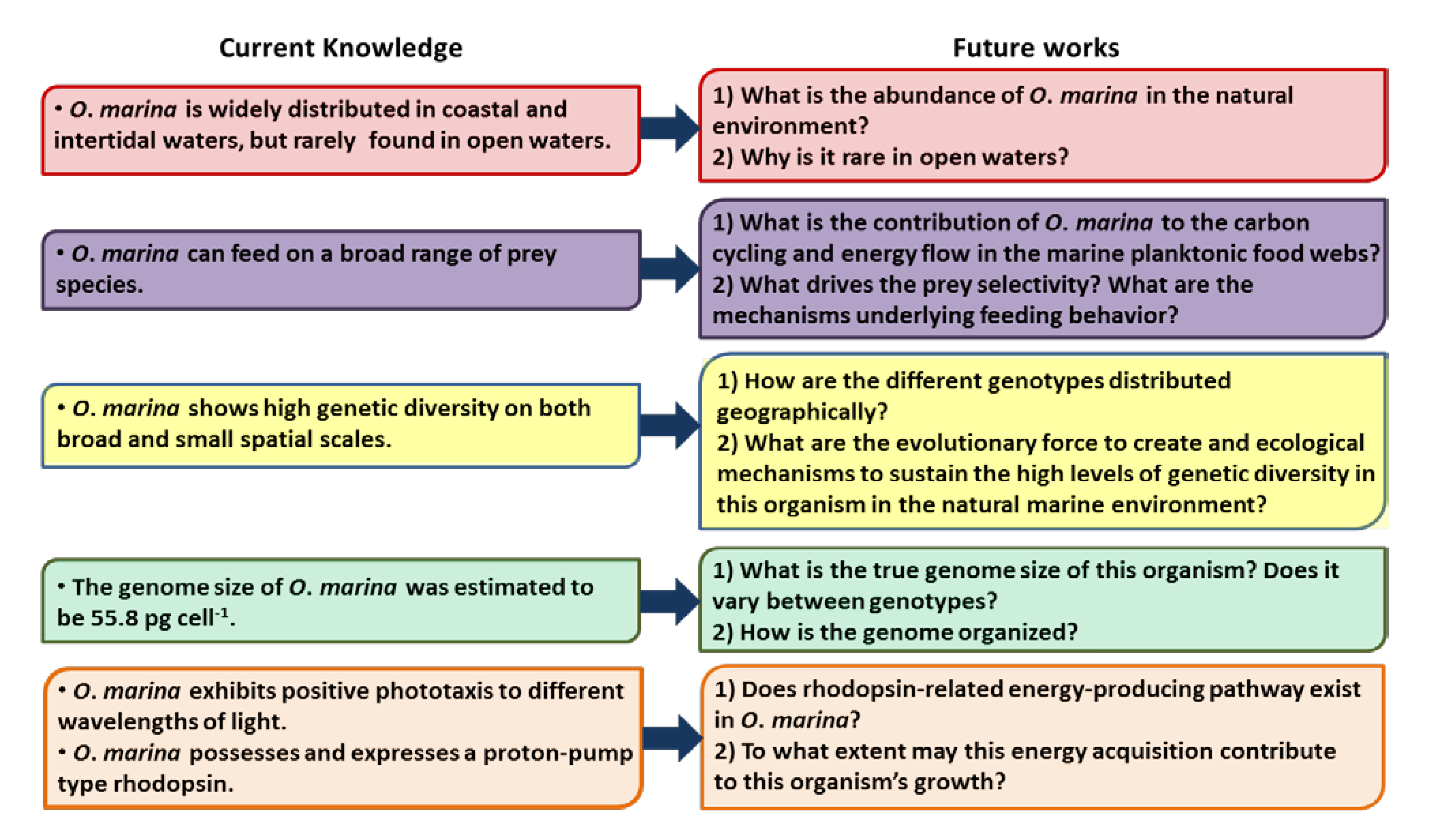
9.1. O. marina Abundance in the Natural Environment
9.2. O. marina Transcriptomic Study
Acknowledgments
Conflict of Interest
References
- Sherr, E.B.; Sherr, B.F. Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 2002, 81, 293–308. [Google Scholar] [CrossRef]
- Roberts, E.C.; Wootton, E.C.; Davidson, K.; Jeong, H.J.; Lowe, C.D.; Montagnes, D.J.S. Feeding in the dinoflagellate Oxyrrhis marina: Linking behaviour with mechanisms. J. Plankton Res. 2011, 33, 603–614. [Google Scholar] [CrossRef]
- Yang, Z.; Jeong, H.J.; Montagnes, D.J.S. The role of Oxyrrhis marina as a model prey: Current work and future directions. J. Plankton Res. 2011, 33, 665–675. [Google Scholar] [CrossRef]
- Lowe, C.D.; Martin, L.E.; Montagnes, D.J.; Watts, P.C. A legacy of contrasting spatial genetic structure on either side of the Atlantic–Mediterranean transition zone in a marine protist. Proc. Natl. Acad. Sci. USA 2012, 109, 20998–21003. [Google Scholar] [CrossRef]
- Jeong, H.J. The ecological roles of heterotrophic dinoflagellates in marine planktonic community. J. Eukaryot. Microbiol. 1999, 46, 190–396. [Google Scholar]
- Sherr, E.B.; Sherr, B.F. Heterotrophic dinoflagellates: A significant component of microzooplankton biomass and major grazers of diatoms in the sea. Mar. Ecol. Prog. Ser. 2007, 352, 187–197. [Google Scholar] [CrossRef]
- Hansen, P.J. Dinophysis—A planktonic dinoflagellate genus which can act both as a prey and a predator of a ciliate. Mar. Ecol. Prog. Ser. 1991, 69, 201–204. [Google Scholar] [CrossRef]
- Jeong, H.J.; Seong, K.A.; Yoo, Y.D.; Kim, T.H.; Kang, N.S.; Kim, S.; Park, J.Y.; Kim, J.S.; Kim, G.H.; Song, J.Y. Feeding and grazing impact by small marine heterotrophic dinoflagellates on heterotrophic bacteria. J. Eukaryot. Microbiol. 2008, 55, 271–288. [Google Scholar] [CrossRef]
- Slamovits, C.H.; Keeling, P.J. Plastid-derived genes in the nonphotosynthetic alveolate Oxyrrhis marina. Mol. Biol. Evol. 2008, 25, 1297–1306. [Google Scholar] [CrossRef]
- Lowe, C.D.; Keeling, P.J.; Martin, L.E.; Slamovits, C.H.; Watts, P.C.; Montagnes, D.J.S. Who is Oxyrrhis marina? Morphological and phylogenetic studies on an unusual dinoflagellate. J. Plankton Res. 2011, 33, 555–567. [Google Scholar] [CrossRef]
- Watts, P.C.; Martin, L.E.; Kimmance, S.A.; Montagnes, D.J.; Lowe, C.D. The distribution of Oxyrrhis marina: A global disperser or poorly characterized endemic? J. Plankton Res. 2011, 33, 579–589. [Google Scholar] [CrossRef]
- Tillmann, U. Phagotrophy by a plastidic haptophyte, Prymnesium patelliferurn. Aquat. Microb. Ecol. 1998, 14, 155–160. [Google Scholar] [CrossRef]
- Tillmann, U. Kill and eat your predator: A winning strategy of the planktonic flagellate Prymnesium parvum. Aquat. Microb. Ecol. 2003, 32, 73–84. [Google Scholar] [CrossRef]
- Suttle, C.A. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Mariani, P.; Botte, V.; Ribera d’Alcalà, M. A numerical investigation of the impact of turbulence on the feeding rates of Oithona davisae. J. Mar. Syst. 2008, 70, 273–286. [Google Scholar] [CrossRef]
- Boakes, D.E.; Codling, E.A.; Thorn, G.J.; Steinke, M. Analysis and modelling of swimming behaviour in Oxyrrhis marina. J. Plankton Res. 2011, 33, 641–649. [Google Scholar] [CrossRef]
- Lowe, C.D.; Day, A.; Kemp, S.J.; Montagnes, D.J. There are high levels of functional and genetic diversity in Oxyrrhis marina. J. Eukaryot. Microbiol. 2005, 52, 250–257. [Google Scholar] [CrossRef]
- Montagnes, D.J.S.; Lowe, C.D.; Martin, L.; Watts, P.C.; Downes-Tettmar, N.; Yang, Z.; Roberts, E.C.; Davidson, K. Oxyrrhis marina growth, sex and reproduction. J. Plankton Res. 2011, 33, 615–627. [Google Scholar] [CrossRef]
- Hartz, A.J.; Sherr, B.F.; Sherr, E.B. Photoresponse in the heterotrophic marine dinoflagellate Oxyrrhis marina. J. Eukaryot. Microbiol. 2011, 58, 171–177. [Google Scholar] [CrossRef]
- Montagnes, D.J.S.; Lowe, C.D.; Roberts, E.C.; Breckels, M.N.; Boakes, D.E.; Davidson, K.; Keeling, P.J.; Slamovits, C.H.; Steinke, M.; Yang, Z.; et al. An introduction to the special issue: Oxyrrhis marina, a model organism? J. Plankton Res. 2011, 33, 549–554. [Google Scholar] [CrossRef]
- Van Meel, L. Etudes hydrobiologiques des eaux saumâtres de belgique: 3. Les étangs galgenweelen à anvers (rive gauche). Bull. K. Belg. Inst. Nat. Wet. 1958, 34, 1–20. (in French). [Google Scholar]
- Scheffel, A. Phaeocystis globosa nov. Spec. Nebst einigen betrachtungen Über die phylogenie niederer, insbesondere brauner organismen. In Wissenschaftliche Meeresuntersuchungen; (in German). Abteilung Helgoland N. F.: Helgoland, Germany, 1900; Volume 4, pp. 1–29. [Google Scholar]
- Conrad, W. Notes protistologiques ix surtroisdinoflagellates de l’eausaumatre. Bull. Mus. Roy. Hist. Nat. Belg. 1939, 15, 1–10. (in French). [Google Scholar]
- Kofoid, C.A.; Swezy, O. The Free-Living Unarmored Dinoflagellata; University of California Press: Berkeley, CA, USA, 1921; Volume 5, p. 538. [Google Scholar]
- Dodge, J.D.; Hart-Jones, B. Marine Dinoflagellates of the British Isles; HMSO: London, UK, 1982. [Google Scholar]
- Cavalier-Smith, T.; Chao, E. Protalveolate phylogeny and systematics and the origins of Sporozoa and dinoflagellates (phylum Myzozoa nom. Nov.). Eur. J. Protistol. 2004, 40, 185–212. [Google Scholar] [CrossRef]
- Lowe, C.D.; Montagnes, D.J.S.; Martin, L.E.; Watts, P.C. Patterns of genetic diversity in the marine heterotrophic flagellate Oxyrrhis marina (alveolata: Dinophyceae). Protist 2010, 161, 212–221. [Google Scholar] [CrossRef]
- Lowe, C.D.; Montagnes, D.J.; Martin, L.E.; Watts, P.C. High genetic diversity and fine-scale spatial structure in the marine flagellate Oxyrrhis marina (Dinophyceae) uncovered by microsatellite loci. PLoS One 2010, 5, e15557. [Google Scholar]
- Lowe, C.D.; Martin, L.E.; Roberts, E.C.; Watts, P.C.; Wootton, E.C.; Montagnes, D.J.S. Collection, isolation, and culturing strategies for Oxyrrhis marina. J. Plankton Res. 2011, 33, 569–578. [Google Scholar] [CrossRef]
- Cachon, J.; Cachon, M.; Salvano, P. The nuclear division of Oxyrrhis marina: An example of the role played by the nuclear envelope in chromosome segregation. Arch. Protistenk. 1979, 122, 43–54. [Google Scholar] [CrossRef]
- Kato, K.H.; Moriyama, A.; Itoh, T.J.; Yamamoto, M.; Horio, T.; Huitorel, P. Dynamic changes in microtubule organization during division of the primitive dinoflagellate Oxyrrhis marina. Biol. Cell 2000, 92, 583–594. [Google Scholar] [CrossRef]
- Saldarriaga, J.F. Multiple protein phylogenies show that Oxyrrhis marina and Perkinsus marinus are early branches of the dinoflagellate lineage. Int. J. Syst. Evol. Microbiol. 2003, 53, 355–365. [Google Scholar] [CrossRef]
- Lenaers, G.; Scholin, C.; Bhaud, Y.; Saint-Hilaire, D.; Herzog, M. A molecular phylogeny of dinoflagellate protists (pyrrhophyta) inferred from the sequence of 24S rRNA divergent domains d1 and d8. J. Mol. Evol. 1991, 32, 53–63. [Google Scholar] [CrossRef]
- Slamovits, C.H.; Saldarriaga, J.F.; Larocque, A.; Keeling, P.J. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J. Mol. Biol. 2007, 372, 356–368. [Google Scholar] [CrossRef]
- Leander, B.S.; Keeling, P.J. Early evolutionary history of dinoflagellates and apicomplexans (alveolata) as inferred from HSP90 and actin phylogenies. J. Phycol. 2004, 40, 341–350. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, S. mRNA editing and spliced-leader RNA trans-splicing groups Oxyrrhis, Noctiluca, Heterocapsa, and Amphidiniumas basal lineages of dinoflagellates. J. Phycol. 2008, 44, 703–711. [Google Scholar] [CrossRef]
- Clarke, K.; Pennick, N. The occurrence of body scales in Oxyrrhis marina dujardin. Br. Phycol. J. 1976, 11, 345–348. [Google Scholar] [CrossRef]
- Clarke, K.; Pennick, N. Flagellar scales in Oxyrrhis marina dujardin. Br. Phycol. J. 1972, 7, 357–360. [Google Scholar] [CrossRef]
- Cachon, M.; Cosson, J.; Cosson, M.P.; Huitorel, P.; Cachon, J. Ultrastructure of the flagellar apparatus of Oxyrrhis marina. Biol. Cell 1988, 63, 159–168. [Google Scholar] [CrossRef]
- Roberts, K.R. The flagellar apparatus of Oxyrrhis marina (pyrrophyta). J. Phycol. 1985, 21, 641–655. [Google Scholar] [CrossRef]
- Roberts, K.; Roberts, J.E. The Flagellar Apparatus and Cytoskeleton of the Dinoflagellates. Protoplasma 1991, 164, 105–122. [Google Scholar] [CrossRef]
- Kato, K.H.; Moriyama, A.; Huitorel, P.; Cosson, J.; Cachon, M.; Sato, H. Isolation of the major basic nuclear protein and its localization on chromosomes of the dinoflagellate Oxyrrhis marina. Biol. Cell 1997, 89, 43–52. [Google Scholar] [CrossRef]
- Zhang, H.; Hou, Y.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar]
- Zhang, H.; Zhuang, Y.; Gill, J.; Lin, S. Proof that dinoflagellate spliced leader (dinosl) is a useful hook for fishing dinoflagellate transcripts from mixed microbial samples: Symbiodinium kawagutii as a case study. Protist 2013, 164, 510–527. [Google Scholar] [CrossRef]
- Hall, R.P. Binary Fission in Oxyrrhis marina Dujardin. University of California Press: Berkeley, CA, USA, 1925; Volume 26, pp. 281–324. [Google Scholar]
- Triemer, R.E. A unique mitotici variation in the marine dinoflagellate Oxyrrhis marina (pyrrophyta). J. Phycol. 1982, 18, 399–411. [Google Scholar] [CrossRef]
- Gao, X.; Li, J. Nuclear division in the marine dinoflagellate Oxyrrhis marina. J. Cell Sci. 1986, 85, 161–175. [Google Scholar]
- Sano, J.; Kato, K.H. Localization and copy number of the protein-codinggenes actin, α-tubulin, and HSP90 in the nucleus of a primitive dinoflagellate, Oxyrrhis marina. Zool. Sci. 2009, 26, 745–753. [Google Scholar] [CrossRef]
- Slamovits, C.H.; Keeling, P.J. Contributions of Oxyrrhis marina to molecular biology, genomics and organelle evolution of dinoflagellates. J. Plankton Res. 2011, 33, 591–602. [Google Scholar] [CrossRef]
- Coats, D.W. Dinoflagellate life-cycle complexities. J. Phycol. 2002, 38, 417–419. [Google Scholar] [CrossRef]
- Santos, S.R.; Coffroth, M.A. Molecular genetic evidence that dinoflagellates belonging to the genus Symbiodinium freudenthal are haploid. Biol. Bull. 2003, 204, 10–20. [Google Scholar]
- Hausmann, K.; Hülsmann, N.; Radek, R. Protistology; E. Schweizerbart’sche Verlagsbuchhandlung: Berlin, Germany, 2003. [Google Scholar]
- Hackett, J.D.; Anderson, D.M.; Erdner, D.L.; Bhattacharya, D. Dinoflagellates: A remarkable evolutionary experiment. Am. J. Bot. 2004, 91, 1523–1534. [Google Scholar] [CrossRef]
- Veldhuis, M.J.W.; Cucci, T.L.; Sieracki, M.E. Cellular DNA content of marine phytoplankton using two new fluorochromes: Taxonomic and ecological implications. J. Phycol. 1997, 33, 527–541. [Google Scholar]
- LaJeunesse, T.C.; Lambert, G.; Anderson, R.A.; Coffroth, M.A.; Galbraith, D.W. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J. Phycol. 2005, 41, 880–886. [Google Scholar]
- Le, Q.; Markovic, P.; Hastings, J.; Jovine, R.; Morse, D. Structure and organization of the peridinin-chlorophyll a-binding protein gene in Gonyaulax polyedra. Mol. Gen. Genet. 1997, 255, 595–604. [Google Scholar]
- Reichman, J.R.; Wilcox, T.P.; Vize, P.D. PCP gene family in Symbiodinium from Hippopus hippopus: Low levels of concerted evolution, isoform diversity, and spectral tuning of chromophores. Mol. Biol. Evol. 2003, 20, 2143–2154. [Google Scholar]
- Bachvaroff, T.R.; Place, A.R. From stop to start: Tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS One 2008, 3, e2929. [Google Scholar] [CrossRef]
- Galluzzi, L.; Bertozzini, E.; Penna, A.; Perini, F.; Garcés, E.; Magnani, M. Analysis of rRNA gene content in the mediterranean dinoflagellate Alexandrium catenella and Alexandrium taylori: Implications for the quantitative real-time PCR-based monitoring methods. J. Appl. Phycol. 2009, 22, 1–9. [Google Scholar]
- Erdner, D.L.; Percy, L.; Keafer, B.; Lewis, J.; Anderson, D.M. A quantitative real-time PCR assay for the identification and enumeration of Alexandrium cysts in marine sediments. Deep Sea Res. Part II Top. Stud. Oceanogr. 2010, 57, 279–287. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, S. Complex gene structure of the form II Rubisco in the dinoflagellate Prorocentrum minimum (Dinophyceae). J. Phycol. 2003, 39, 1160–1171. [Google Scholar] [CrossRef]
- Li, L.; Hastings, J.W. The structure and organization of the luciferase gene in the photosynthetic dinoflagellate Gonyaulax polyedra. Plant Mol. Biol. 1998, 36, 275–284. [Google Scholar]
- Zhang, H.; Hou, Y.; Lin, S. Isolation and characterization of proliferating cell nuclear antigen from the dinoflagellate Pfiesteria piscicida. J. Eukaryot. Microbiol. 2006, 53, 142–150. [Google Scholar] [CrossRef]
- Zhang, H.; Dungan, C.F.; Lin, S. Introns, alternative splicing, spliced leader trans-splicing and differential expression of pcna and cyclin in Perkinsus marinus. Protist 2011, 162, 154–167. [Google Scholar]
- Beauchemin, M.; Roy, S.; Daoust, P.; Dagenais-Bellefeuille, S.; Bertomeu, T.; Letourneau, L.; Lang, B.F.; Morse, D. Dinoflagellate tandem array gene transcripts are highly conserved and not polycistronic. Proc. Natl. Acad. Sci. USA 2012, 109, 15793–15798. [Google Scholar]
- Lowe, C.D.; Mello, L.V.; Samatar, N.; Martin, L.E.; Montagnes, D.J.S.; Watts, P.C. The transcriptome of the novel dinoflagellate Oxyrrhis marina (alveolata: Dinophyceae): Response to salinity examined by 454 sequencing. BMC Genomics 2011. [Google Scholar] [CrossRef]
- Waller, R.F.; Slamovits, C.H.; Keeling, P.J. Lateral gene transfer of a multigene region from cyanobacteria to dinoflagellates resulting in a novel plastid-targeted fusion protein. Mol. Biol. Evol. 2006, 23, 1437–1443. [Google Scholar]
- Delwiche, C.F.; Palmer, J.D. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 1996, 13, 873–882. [Google Scholar] [CrossRef]
- Morse, D.; Salois, P.; Markovic, P.; Hastings, J.W. A nuclear-encoded form II rubisco in dinoflagellates. Science 1995, 268, 1622–1624. [Google Scholar]
- Lin, S.; Zhang, H.; Spencer, D.F.; Norman, J.E.; Gray, M.W. Widespread and extensive editing of mitochondrial mRNAs in dinoflagellates. J. Mol. Biol. 2002, 320, 727–739. [Google Scholar]
- Jackson, C.J.; Norman, J.E.; Schnare, M.N.; Gray, M.W.; Keeling, P.J.; Waller, R.F. Broad genomic and transcriptional analysis reveals a highly derived genome in dinoflagellate mitochondria. BMC Biol. 2007. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, S. Mitochondrial cytochrome b mRNA editing in dinoflagellates: Possible ecological and evolutionary associations? J. Eukaryot. Microbiol. 2005, 52, 538–545. [Google Scholar] [CrossRef]
- Zhang, H.; Bhattacharya, D.; Lin, S. A three-gene dinoflagellate phylogeny suggests monophyly of prorocentrales and a basal position for Amphidinium and Heterocapsa. J. Mol. Evol. 2007, 65, 463–474. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Lippmeier, J.C.; Apt, K.E.; Delwiche, C.F. Plastid genes in a non-photosynthetic dinoflagellate. Protist 2007, 158, 105–117. [Google Scholar]
- Lin, S.; Zhang, H.; Gray, M.W. RNA Editing in Dinoflagellates and Its Implications for the Evolutionary History of the Editing Machinery. In RNA and DNA Editing: Molecular Mechanisms and Their Integration into Biological Systems; Smith, H.C., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; pp. 280–309. [Google Scholar]
- Jackson, C.; Gornik, S.; Waller, R. The mitochondrial genome and transcriptome of the basal dinoflagellate Hematodinium sp.: Character evolution within the highly derived mitochondrial genomes of dinoflagellates. Genome Biol. Evol. 2012, 4, 59–72. [Google Scholar] [CrossRef]
- Williamson, D.H.; Gardner, M.J.; Preiser, P.; Moore, D.J.; Rangachari, K.; Wilson, R.J. The evolutionary origin of the 35 kb circular DNA of Plasmodium falciparum: New evidence supports a possible rhodophyte ancestry. Mol. Gen. Genet. 1994, 243, 249–252. [Google Scholar]
- Denny, P.W.; Preiser, P.R.; Rangachari, K.; Roberts, K.; Roy, A.; Whyte, A.; Strath, M.; Moore, D.J.; Moore, P.W.; Williamson, D.H. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 1996, 261, 155–172. [Google Scholar] [CrossRef]
- Huang, J.; Mullapudi, N.; Sicheritz-Ponten, T.; Kissinger, J.C. A first glimpse into the pattern and scale of gene transfer in the apicomplexa. Int. J. Parasitol. 2004, 34, 265–274. [Google Scholar] [CrossRef]
- Archibald, J.M. Plastid evolution: Remnant algal genes in ciliates. Curr. Biol. 2008, 18, R663–R665. [Google Scholar] [CrossRef]
- Reyes-Prieto, A.; Moustafa, A.; Bhattacharya, D. Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic. Curr. Biol. 2008, 18, 956–962. [Google Scholar] [CrossRef]
- Stelter, K.; El-Sayed, N.M.; Seeber, F. The expression of a plant-type ferredoxin redox system provides molecular evidence for a plastid in the early dinoflagellate Perkinsus marinus. Protist 2007, 158, 119–130. [Google Scholar] [CrossRef]
- Matsuzaki, M.; Kuroiwa, H.; Kuroiwa, T.; Kita, K.; Nozaki, H. A cryptic algal group unveiled: A plastid biosynthesis pathway in the oyster parasite Perkinsus marinus. Mol. Biol. Evol. 2008, 25, 1167–1179. [Google Scholar] [CrossRef]
- Sheiner, L.; Vaidya, A.B.; McFadden, G.I. The metabolic roles of the endosymbiotic organelles of Toxoplasma and Plasmodium spp. Curr. Opin. Microbiol. 2013, 16, 452–458. [Google Scholar] [CrossRef]
- Cavalier-smith, T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree 1,2. J. Eukaryot. Microbiol. 1999, 46, 347–366. [Google Scholar] [CrossRef]
- Patron, N.J.; Rogers, M.B.; Keeling, P.J. Gene replacement of fructose-1,6-bisphosphate aldolase supports the hypothesis of a single photosynthetic ancestor of chromalveolates. Eukaryot. Cell 2004, 3, 1169–1175. [Google Scholar] [CrossRef]
- Keeling, P.J. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot. Microbiol. 2009, 56, 1–8. [Google Scholar] [CrossRef]
- Goldman, J.C.; Dennett, M.R.; Gordin, H. Dynamics of herbivorous grazing by the heterotrophic dinoflagellate Oxyrrhis marina. J. Plankton Res. 1989, 11, 391–407. [Google Scholar] [CrossRef]
- Hansen, F.C.; Witte, H.J.; Passarge, J. Grazing in the heterotrophic dinoflagellate Oxyrrhis marina: Size selectivity and preference for calcified Emiliania huxleyi cells. Aquat. Microb. Ecol. 1996, 10, 307–313. [Google Scholar] [CrossRef]
- Jeong, H.J.; Kang, H.; Shim, J.H.; Park, J.K.; Kim, J.S.; Song, J.Y.; Choi, H.J. Interactions among the toxic dinoflagellate Amphidinium carterae, the heterotrophic dinoflagellate Oxyrrhis marina, and the calanoid copepods acartia spp. Mar. Ecol. Prog. Ser. 2001, 218, 77–86. [Google Scholar] [CrossRef]
- Jeong, H.J.; Kim, J.S.; Yoo, Y.D.; Kim, S.T.; Kim, T.H.; Park, M.G.; Lee, C.H.; Seong, K.A.; Rang, N.S.; Shim, J.H. Feeding by the heterotrophic dinoflagellate Oxyrrhis marina on the red-tide raphidophyte Heterosigma akashiwo: A potential biological method to control red tides using mass-cultured grazers. J. Eukaryot. Microbiol. 2003, 50, 274–282. [Google Scholar] [CrossRef]
- Hammer, A.; Grüttner, C.; Schumann, R. The effect of electrostatic charge of food particles on capture efficiency by Oxyrrhis marina dujardin (dinoflagellate). Protist 1999, 150, 375–382. [Google Scholar] [CrossRef]
- Hammer, A.; Gruttner, C.; Schumann, R. New biocompatible tracer particles: Use for estimation of microzooplankton grazing, digestion, and growth rates. Aquat. Microb. Ecol. 2001, 24, 153–161. [Google Scholar] [CrossRef]
- Wootton, E.C.; Zubkov, M.V.; Jones, D.H.; Jones, R.H.; Martel, C.M.; Thornton, C.A.; Roberts, E.C. Biochemical prey recognition by planktonic protozoa. Environ. Microbiol. 2007, 9, 216–222. [Google Scholar] [CrossRef]
- Sieburth, J.M. Acrylic acid, an “antibiotic” principle in phaeocystis blooms in antarctic waters. Science 1960, 132, 676–677. [Google Scholar]
- Barlow, R.; Burkill, P.; Mantoura, R. Grazing and degradation of algal pigments by marine protozoan Oxyrrhis marina. J. Exp. Mar. Biol. Ecol. 1988, 119, 119–129. [Google Scholar] [CrossRef]
- Flynn, K.J.; Davidson, K.; Cunningham, A. Prey selection and rejection by a microflagellate: Implications for the study and operation of microbial food webs. J. Exp. Mar. Biol. Ecol. 1996, 196, 357–372. [Google Scholar] [CrossRef]
- Monger, B.C.; Landry, M.R.; Brown, S.L. Feeding selection of heterotrophic marine nanoflagellates based on the surface hydrophobicity of their picoplankton prey. Limnol. Oceanogr. 1999, 44, 1917–1927. [Google Scholar] [CrossRef]
- John, E.; Davidson, K. Prey selectivity and the influence of prey carbon: Nitrogen ratio on microflagellate grazing. J. Exp. Mar. Biol. Ecol. 2001, 260, 93–111. [Google Scholar] [CrossRef]
- Matz, C.; Jurgens, K. Effects of hydrophobic and electrostatic cell surface properties of bacteria on feeding rates of heterotrophic nanoflagellates. Appl. Environ. Microbiol. 2001, 67, 814–820. [Google Scholar] [CrossRef]
- Matz, C.; Jürgens, K. High motility reduces grazing mortality of planktonic bacteria. Appl. Environ. Microbiol. 2005, 71, 921–929. [Google Scholar] [CrossRef]
- Matz, C.; Boenigk, J.; Arndt, H.; Jürgens, K. Role of bacterial phenotypic traits in selective feeding of the heterotrophic nanoflagellate Spumella sp. Aquat. Microb. Ecol. 2002, 27, 137–148. [Google Scholar] [CrossRef]
- Wolfe, G.V.; Steinke, M.; Kirst, G.O. Grazing-activated chemical defence in a unicellular marine alga. Nature 1997, 387, 894–897. [Google Scholar]
- Evans, C.; Wilson, W.H. Preferential grazing of Oxyrrhis marina on virus-infected Emiliania huxleyi. Limnol. Oceanogr. 2008, 53, 2035–2040. [Google Scholar] [CrossRef]
- Droop, M.R. Nutritional investigation of phagotrophic protozoa under axenic conditions. Helgoländer Wiss. Meeresunters. 1970, 20, 272–277. [Google Scholar]
- Droop, M.R. Water-soluble factors in the nutrition of Oxyrrhis marina. J. Mar. Biol. Assoc. UK 1959, 38, 605–620. [Google Scholar] [CrossRef]
- Droop, M.R.; Pennock, J.F. Terpenoid quinones and steroids in the nutrition of Oxyrrhis marina. J. Mar. Biol. Assoc. UK 1971, 51, 455–470. [Google Scholar] [CrossRef]
- Öpik, H.; Flynn, K. The digestive process of the dinoflagellate, Oxyrrhis marina dujardin, feeding on the chlorophyte, Dunaliella primolecta butcher: A combined study of ultrastructure and free amino acids. New Phytol. 1989, 113, 143–151. [Google Scholar] [CrossRef]
- Mast, S.; Stahler, N. The relation between luminous intensity, adaptation to light, and rate of locomotion in Amoeba proteus (leidy). Biol. Bull. 1937, 73, 126–133. [Google Scholar] [CrossRef]
- Podesta, A.; Marangoni, R.; Vilani, C.; Colombetti, G. A rhodopsin-like molecule on the plasma membrane of Fabrea salina. J. Eukaryot. Microbiol. 1994, 41, 565–569. [Google Scholar] [CrossRef]
- Seibach, M.; Hader, D.P.; Kuhlmann, H.W. Phototaxis in Chlamydodon mnemosyne: Determination of the illuminance-response curve and the action spectrum. J. Photochem. Photobiol. B Biol. 1999, 49, 35–40. [Google Scholar] [CrossRef]
- Cadetti, L.; Marroni, F.; Marangoni, R.; Kuhlmann, H.-W.; Gioffre, D.; Colombetti, G. Phototaxis in the ciliated protozoan Ophryoglena flava: Dose-effect curves and action spectrum determination. J. Photochem. Photobiol. B Biol. 2000, 57, 41–50. [Google Scholar] [CrossRef]
- Saranak, J.; Foster, K.W. Photoreceptor for curling behavior in Peranema trichophorum and evolution of eukaryotic rhodopsins. Eukaryot. Cell 2005, 4, 1605–1612. [Google Scholar] [CrossRef]
- Lobban, C.S.; Hallam, S.J.; Mukherjee, P.; Petrich, J.W. Photophysics and multifunctionality of hypericin-like pigments in heterotrich ciliates: A phylogenetic perspective. Photochem. Photobiol. 2007, 83, 1074–1094. [Google Scholar] [CrossRef]
- Fabczak, H.; Sobierajska, K.; Fabczak, S. A rhodopsin immunoanalog in the related photosensitive protozoans Blepharisma japonicum and Stentor coeruleus. Photochem. Photobiol. Sci. 2008, 7, 1041–1045. [Google Scholar]
- Jakobsen, H.H.; Strom, S.L. Circadian cycles in growth and feeding rates of heterotrophic protist plankton. Limnol. Oceanogr. 2004, 49, 1915–1922. [Google Scholar] [CrossRef]
- Strom, S.L. Light-aided digestion, grazing and growth in herbivorous protists. Aquat. Microb. Ecol. 2001, 23, 253–261. [Google Scholar] [CrossRef]
- Skovgaard, A. A phagotrophically derivable growth factor in the plastic dinoflagellates Gyrodinium resplendens. J. Phycol. 2000, 36, 1069–1078. [Google Scholar] [CrossRef]
- Jakobsen, H.H.; Hansen, P.J.; Larsen, J. Growth and grazing responses of two chloroplast-retaining dinoflagellates: Effect of irradiance and prey species. Mar. Ecol. Prog. Ser. 2000, 201, 121–128. [Google Scholar] [CrossRef]
- Li, A.; Stoecker, D.K.; Adolf, J.E. Feeding, pigmentation, photosynthesis and growth of the mixotrophic dinoflagellate Gyrodinium galatheanum. Aquat. Microb. Ecol. 1999, 19, 163–176. [Google Scholar] [CrossRef]
- Moran, M.A.; Zepp, R.G. Role of photoreactions in the formation of biologically labile compounds from dissolved organic matter. Limnol. Oceanogr. 1997, 42, 1307–1316. [Google Scholar] [CrossRef]
- Klein, B.; Gieskes, W.W.; Krray, G.G. Digestion of chlorophylls and carotenoids by the marine protozoan Oxyrrhis marina studied by HPLC analysis of algal pigments. J. Plankton Res. 1986, 8, 827–836. [Google Scholar] [CrossRef]
- Béja, O.; Aravind, L.; Koonin, E.V.; Suzuki, M.T.; Hadd, A.; Nguyen, L.P.; Jovanovich, S.B.; Gates, C.M.; Feldman, R.A.; Spudich, J.L. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science 2000, 289, 1902–1906. [Google Scholar] [CrossRef]
- Béja, O.; Spudich, E.N.; Spudich, J.L.; Leclerc, M.; DeLong, E.F. Proteorhodopsin phototrophy in the ocean. Nature 2001, 411, 786–789. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, H.; Zhuang, Y.; Tran, B.; Gill, J. Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc. Natl. Acad. Sci. USA 2010, 107, 20033–20038. [Google Scholar] [CrossRef]
- Slamovits, C.H.; Okamoto, N.; Burri, L.; James, E.R.; Keeling, P.J. A bacterial proteorhodopsin proton pump in marine eukaryotes. Nat. Commun. 2011. [Google Scholar] [CrossRef]
- Kent, W.S. A Manual of the Infusoria: Including a Description of All Known Flagellate, Ciliate, and Tentaculiferous Protozoa, British and Foreign, and an Account of the Organization and the Affinities of the Sponges; D. Bogue: London, UK, 1880. [Google Scholar]
- Senn, G. Oxyrrhis, Nephroselmis und einige Euflagellaten, nebst Bemerkungen uber deren System; Wilhelm Engelmann: Leipzig, Germany, 1911; pp. 604–672. [Google Scholar]
- Cosson, J.; Cachon, M.; Cachon, J.; Cosson, M.-P. Swimming behaviour of the unicellular biflagellate Oxyrrhis marina: In vivo and in vitro movement of the two flagella. Biol. Cell 1988, 63, 117–126. [Google Scholar] [CrossRef]
- Bartumeus, F.; Peters, F.; Pueyo, S.; Marrasé, C.; Catalan, J. Helical lévy walks: Adjusting searching statistics to resource availability in microzooplankton. Proc. Natl. Acad. Sci. USA 2003, 100, 12771–12775. [Google Scholar] [CrossRef]
- Crenshaw, H.C. A new look at locomotion in microorganisms: Rotating and translating. Am. Zool. 1996, 36, 608–618. [Google Scholar]
- Tarran, G.A. Aspects of Grazing Behaviour of the Marine Dinoflagellate Oxyrrhis marina, Dujardin; University of Southampton: Southampton, UK, 1991. [Google Scholar]
- Menden-Deuer, S.; Grünbaum, D. Individual foraging behaviors and population distributions of a planktonic predator aggregating to phytoplankton thin layers. Limnol. Oceanogr. 2006, 51, 109–116. [Google Scholar] [CrossRef]
- Grünbaum, D. Predicting availability to consumers of spatially and temporally variable resources. Hydrobiologia 2002, 480, 175–191. [Google Scholar] [CrossRef]
- Hartz, A.J.; Sherr, B.F.; Sherr, E.B. Using inhibitors to investigate the involvement of cell signaling in predation by marine phagotrophic protists. J. Eukaryot. Microbiol. 2008, 55, 18–21. [Google Scholar] [CrossRef]
- Fenchel, T. Ecology of Protozoa: The Biology of Free-Living Phagotrophic Protists; Springer-Verlag: Madison, WI, USA, 1987. [Google Scholar]
- Höhfeld, I.; Melkonian, M. Lifting the curtain? The microtubular cytoskeleton of Oxyrrhis marina (Dinophyceae) and its rearrangement during phagocytosis. Protist 1998, 149, 75–88. [Google Scholar] [CrossRef]
- Barker, H.A. The culture and physiology of the marine dinoflagellates. Arch. Microbiol. 1935, 6, 157–181. [Google Scholar]
- Flynn, K.J.; Davidson, K. Predator-prey interactions between Isochrysis galbana and Oxyrrhis marina. II. Release of non-protein amines and faeces during predation of isochrysis. J. Plankton Res. 1993, 15, 893–905. [Google Scholar] [CrossRef]
- Bretler, W. Continuous breeding of marine pelagic copepods in the presence of heterotrophic dinoflagellates. Mar. Ecol. Prog. Ser. 1980, 2, 229–233. [Google Scholar] [CrossRef]
- Hansen, B.; Bjørnsen, P.K.; Hansen, P.J. The size ratio between planktonic predators and their prey. Limnol. Oceanogr. 1994, 39, 395–403. [Google Scholar] [CrossRef]
- Breteler, W.K.; Schogt, N.; Baas, M.; Schouten, S.; Kraay, G. Trophic upgrading of food quality by protozoans enhancing copepod growth: Role of essential lipids. Mar. Biol. 1999, 135, 191–198. [Google Scholar] [CrossRef]
- Scott, J. Further nutritional studies on the marine rotifer encentrum linnhei. Rotifer Symp. IV 1987, 42, 303–306. [Google Scholar] [CrossRef]
- Pfiester, L.A.; Anderson, D.M. Dinoflagellate reproduction. In The Biology of Dinoflagellates; Taylor, F.J.R., Ed.; Blackwell Scientific Publications: Hoboken, NJ, USA, 1987; Volume 21, pp. 611–648. [Google Scholar]
- Whiteley, A.; Burkill, P.; Sleigh, M. Rapid method for cell cycle analysis in a predatory marine dinoflagellate. Cytometry 1993, 14, 909–915. [Google Scholar] [CrossRef]
- Lin, S.; Mulholland, M.R.; Zhang, H.; Feinstein, T.N.; Jochem, F.J.; Carpenter, E.J. Intense grazing and prey-dependent growth of Pfiesteria piscicida (Dinophyceae). J. Phycol. 2004, 40, 1062–1073. [Google Scholar] [CrossRef]
- Begun, A.; Orlova, T.Y.; Selina, M. A “bloom” in the water of Amursky bay (sea of Japan) caused by the dinoflagellate Oxyrrhis marina dujardin, 1841. Russ. J. Mar. Biol. 2004, 30, 51–55. [Google Scholar] [CrossRef]
- Kimmance, S.A.; Atkinson, D.; Montagnes, D.J. Do temperature-food interactions matter? Responses of production and its components in the model heterotrophic flagellate Oxyrrhis marina. Aquat. Microb. Ecol. 2006, 42, 63–73. [Google Scholar] [CrossRef]
- Jonsson, P.R. Tidal rhythm of cyst formation in the rock pool ciliate Strombidium oculatum gruber (ciliophora, oligotrichida): A description of the functional biology and an analysis of the tidal synchronization of encystment. J. Exp. Mar. Biol. Ecol. 1994, 175, 77–103. [Google Scholar] [CrossRef]
- Anderson, D.M.; Wall, D. Potential importance of benthic cysts of Gonyaulax tamarensis and G. excavata in initiating toxic dinoflagellate blooms. J. Phycol. 1978, 14, 224–234. [Google Scholar] [CrossRef]
- Matthiessen, J.; de Vernal, A.; Head, M.; Okolodkov, Y.; Harland, R. Modern organic-walled dinoflagellate cysts in Arctic marine environments and their (paleo-) environmental significance. Paläontologische Zeitschrift 2005, 79, 3–51. [Google Scholar]
- Vink, A.; Zonneveld, K.A.F.; Willems, H. Organic-walled dinoflagellate cysts in western equatorial Atlantic surface sediments: Distribution and their relation to environment. Rev. Palaeobot. Palynol. 2000, 112, 247–286. [Google Scholar] [CrossRef]
- Wall, D. Biological problems concerning fossilizable dinoflagellates. Geosci. Man 1971, 2, 1–15. [Google Scholar]
- Droop, M.R. A note on some physical conditions for cultivating Oxyrrhis marina. J. Mar. Biol. Assoc. UK 1959, 38, 599–604. [Google Scholar] [CrossRef]
- Havskum, H. Effects of small-scale turbulence on interactions between the heterotrophic dinoflagellate Oxyrrhis marina and its prey, Isochrysis sp. Ophelia 2003, 57, 125–135. [Google Scholar] [CrossRef]
- Pedersen, M.F.; Hansen, P.J. Effects of high pH on the growth and survival of six marine heterotrophic protists. Mar. Ecol. Prog. Ser. 2003, 260, 33–41. [Google Scholar] [CrossRef]
- Peters, F.; Marrasé, C. Effects of turbulence on plankton: An overview of experimental evidence and some theoretical considerations. Mar. Ecol. Prog. Ser. 2000, 205, 291–306. [Google Scholar] [CrossRef]
- Davidson, K.; Sayegh, F.; Montagnes, D.J.S. Oxyrrhis marina-based models as a tool to interpret protozoan population dynamics. J. Plankton Res. 2011, 33, 651–663. [Google Scholar] [CrossRef]
- Johnson, M. Physical control of plankton population abundance and dynamics in intertidal rock pools. Hydrobiologia 2000, 440, 145–152. [Google Scholar] [CrossRef]
- Hansson, H.G. South scandinavian marine protoctista. Provisional Check-list Compiled at the Tjarno Marine Biological Laboratory. Available online: http://www.yumpu.com/it/document/view/5925990/south-scandinavian-marine-protoctista-protoctista-tmbl (accessed on 10 October 2013).
- Orlova, T.Y.; Stonik, I.; Shevchenko, O. Flora of planktonic microalgae of Amursky bay, sea of Japan. Russ. J. Mar. Biol. 2009, 35, 60–78. [Google Scholar] [CrossRef]
- Quevedo, M.; Anado, R. Spring microzooplankton composition, biomass and potential grazing in the central cantabrian coast (southern bay of biscay). Oceanol. Acta 2000, 23, 297–310. [Google Scholar] [CrossRef]
- Johnson, M.D.; Rome, M.; Stoecker, D.K. Microzooplankton grazing on Prorocentrum minimum and Karlodinium micrum in chesapeake bay. Limnol. Oceanogr. 2003, 48, 238–248. [Google Scholar] [CrossRef]
- Galluzzi, L.; Penna, A.; Bertozzini, E.; Vila, M.; Garces, E.; Magnani, M. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 2004, 70, 1199–1206. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, S. Development of a cob-18S rRNA gene real-time PCR assay for quantifying Pfiesteria shumwayae in the natural environment. Appl. Environ. Microbiol. 2005, 71, 7053–7063. [Google Scholar] [CrossRef]
- Touzet, N.; Keady, E.; Raine, R.; Maher, M. Evaluation of taxa-specific real-time PCR, whole-cell fish and morphotaxonomy analyses for the detection and quantification of the toxic microalgae Alexandrium minutum (Dinophyceae), global clade ribotype. FEMS Microbiol. Ecol. 2009, 67, 329–341. [Google Scholar] [CrossRef]
- Dyhrman, S.T.; Erdner, D.; Du, J.L.; Galac, M.; Anderson, D.M. Molecular quantification of toxic Alexandrium fundyense in the gulf of maine using real-time PCR. Harmful Algae 2006, 5, 242–250. [Google Scholar] [CrossRef]
- Hosoi-Tanabe, S.; Sako, Y. Species-specific detection and quantification of toxic marine dinoflagellates Alexandrium tamarense and A. catenella by real-time PCR assay. Mar. Biotechnol. 2005, 7, 506–514. [Google Scholar] [CrossRef]
- Wang, L.; Zhuang, Y.; Zhang, H.; Lin, X.; Lin, S. DNA barcoding species in Alexandrium tamarense complex using ITS and proposing designation of five species. Harmful Algae 2013, in press. [Google Scholar]
- Yuan, J.; Mi, T.; Zhen, Y.; Yu, Z. Development of a rapid detection and quantification method of Karenia mikimotoi by real-time quantitative PCR. Harmful Algae 2012, 17, 83–91. [Google Scholar] [CrossRef]
- Zhang, H.; Litaker, W.; Vandersea, M.W.; Tester, P.; Lin, S. Geographic distribution of Karlodinium veneficum in the US east coast as detected by ITS-ferredoxin real-time PCR assay. J. Plankton Res. 2008, 30, 905–922. [Google Scholar] [CrossRef]
- Bowers, H.A.; Tengs, T.; Glasgow, H.B.; Burkholder, J.M.; Rublee, P.A.; Oldach, D.W. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 2000, 66, 4641–4648. [Google Scholar] [CrossRef]
- Mieog, J.C.; van Oppen, M.J.; Berkelmans, R.; Stam, W.T.; Olsen, J.L. Quantification of algal endosymbionts (Symbiodinium) in coral tissue using real-time PCR. Mol. Ecol. Resour. 2009, 9, 74–82. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Guo, Z.; Zhang, H.; Liu, S.; Lin, S. Biology of the Marine Heterotrophic Dinoflagellate Oxyrrhis marina: Current Status and Future Directions. Microorganisms 2013, 1, 33-57. https://doi.org/10.3390/microorganisms1010033
Guo Z, Zhang H, Liu S, Lin S. Biology of the Marine Heterotrophic Dinoflagellate Oxyrrhis marina: Current Status and Future Directions. Microorganisms. 2013; 1(1):33-57. https://doi.org/10.3390/microorganisms1010033
Chicago/Turabian StyleGuo, Zhiling, Huan Zhang, Sheng Liu, and Senjie Lin. 2013. "Biology of the Marine Heterotrophic Dinoflagellate Oxyrrhis marina: Current Status and Future Directions" Microorganisms 1, no. 1: 33-57. https://doi.org/10.3390/microorganisms1010033




