Flavin-Dependent Redox Transfers by the Two-Component Diketocamphane Monooxygenases of Camphor-Grown Pseudomonas putida NCIMB 10007
Abstract
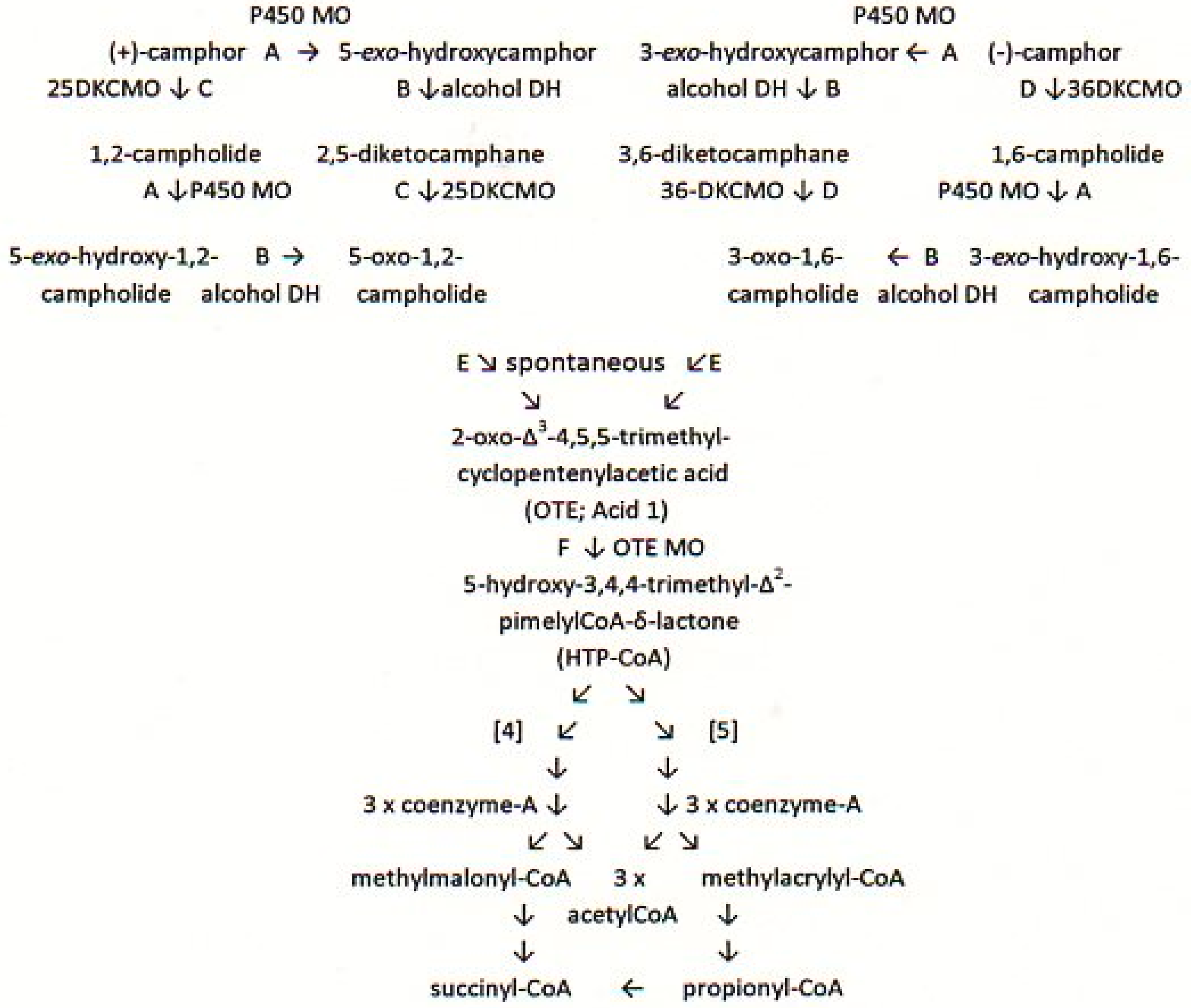
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Maintenance, and Growth Conditions
2.2. Purification of PdR, Pdx, 2,5- , and 3,6-DKCMO
2.3. Purification of FMN-Reductase Activities
2.4. Kinetic Studies of Frp1, Frp2, Fred and PdR
2.5. Reaction Mechanism of Frp1, Frp2 and Fred
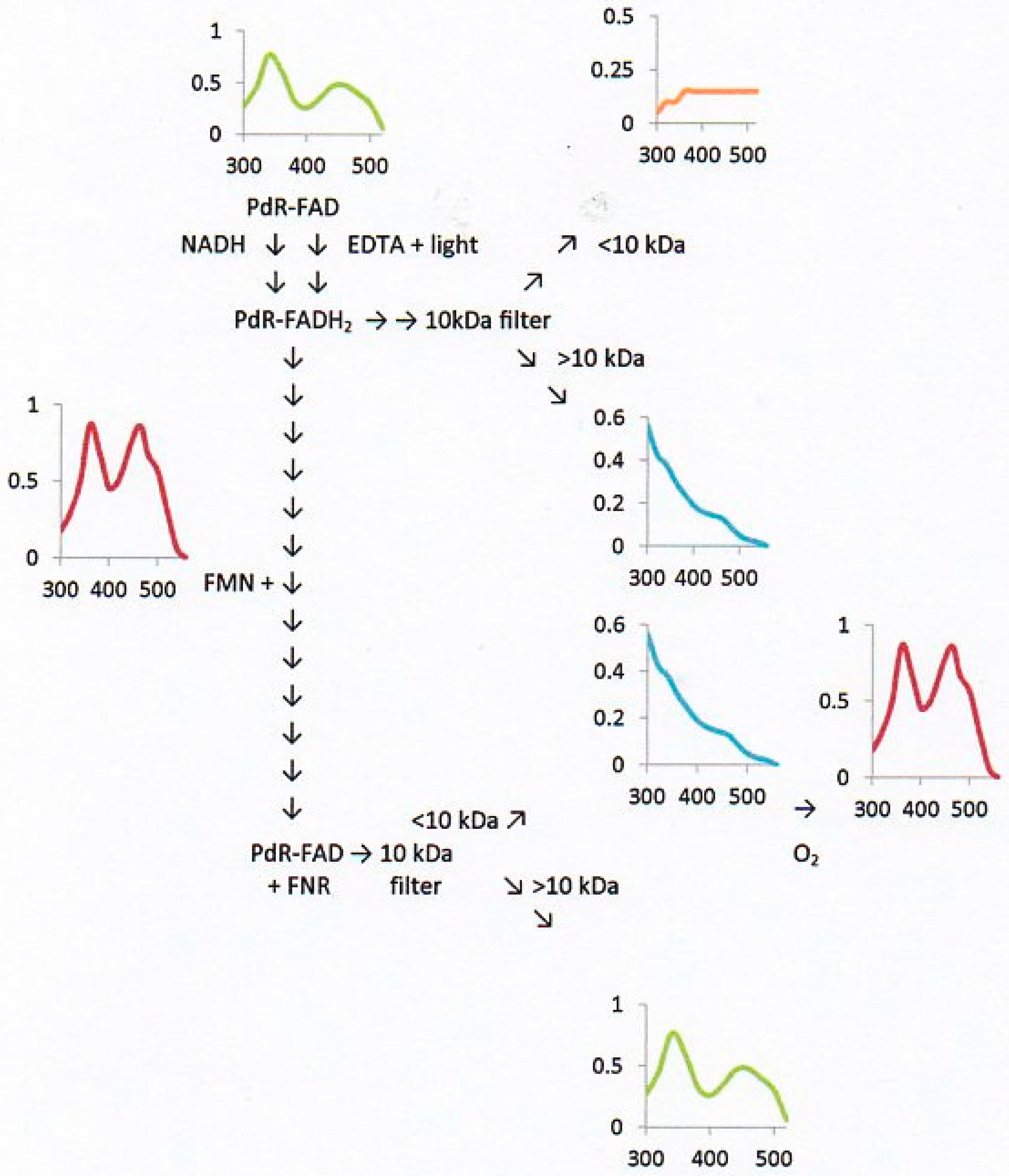
2.6. Flavin Transfer by PdR
2.7. Flavin Binding to the DKCMOs and Kd Values for FMN and FNR
2.8. Biocatalytic Reactions with Combinations of Purified Enzymes
2.9. Reproducibility
2.10. Chemicals and General Procedures
3. Results
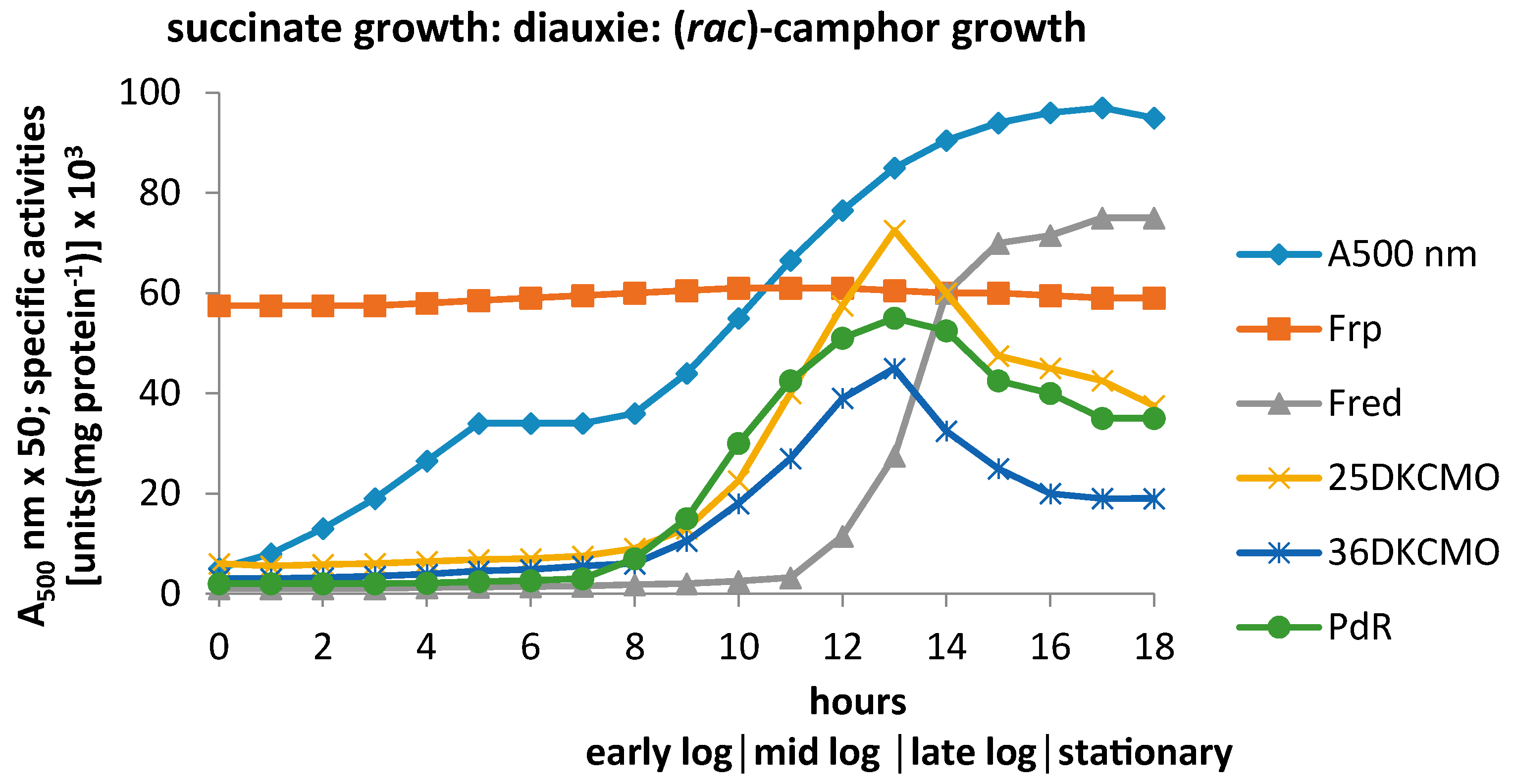
3.1. Specific Activities of Key Enzymes Involved in FNR Generation and Deployment throughout Growth of P. putida NCIMB 10007 on Camphor
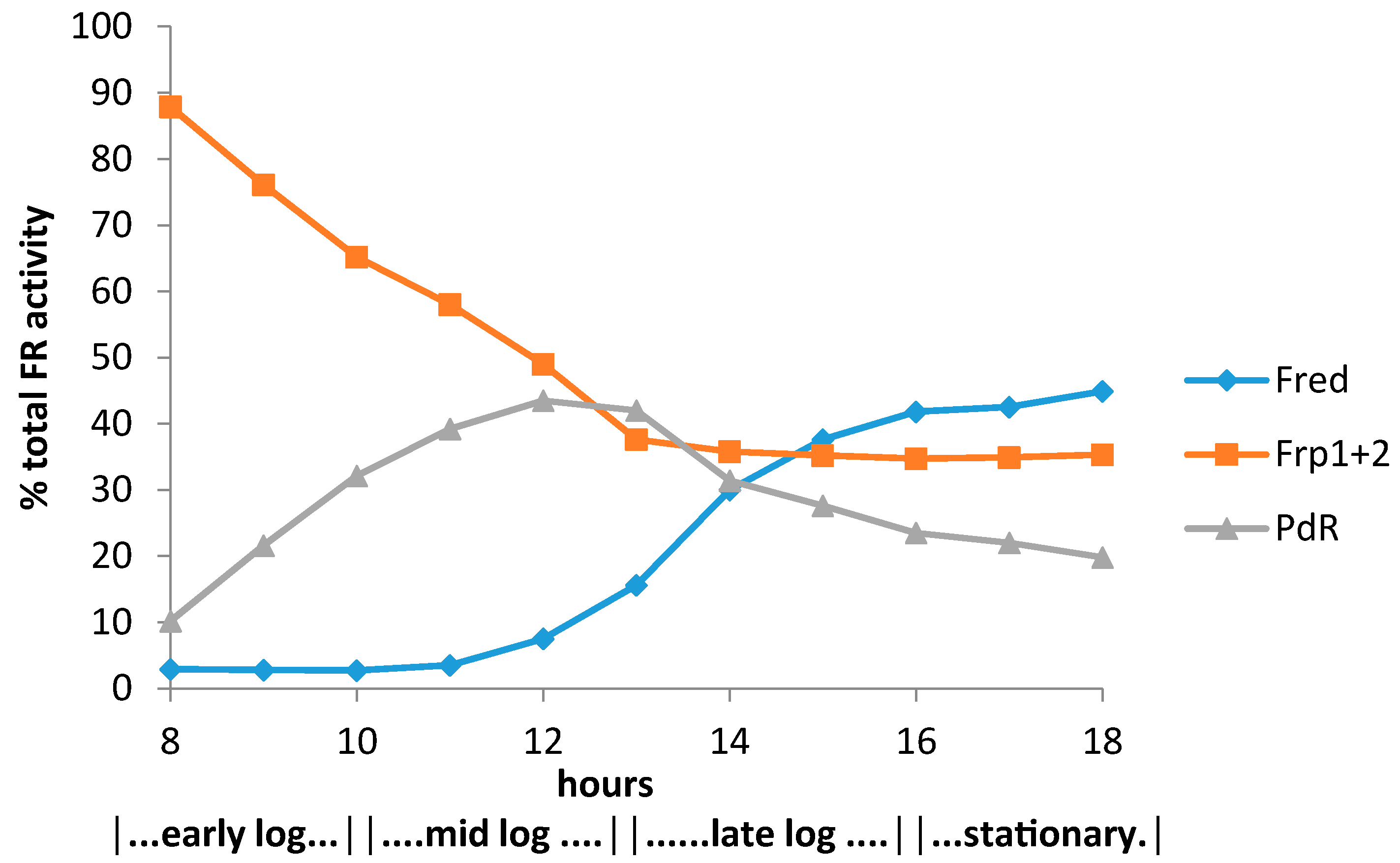
3.2. Relevant Kinetic Data for the Alternative FNR-Generating Activities in Camphor-Grown P. putida NCIMB 10007
3.2.1. Constitutive Low MW FNR-Generating Activity: Ferric Reductases
3.2.2. Inducible Low MW FNR-Generating Activity: Fred
3.2.3. Inducible High MW FRs; PdR
3.3. Relevant Kinetic Data for the FNR-Dependent Isoenzymic DKCMO Activities in Camphor—Grown P. putida NCIMB 10007
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gunsalus, I.C. Early reactions in the degradation of camphor: P450cam hydroxylase. In Degradation of Synthetic Organic Molecules in the Biosphere; National Academy of Sciences: Washington DC, USA, 1972; pp. 137–146. [Google Scholar]
- Bradshaw, W.H.; Conrad, H.E.; Corey, E.J.; Gunsalus, I.C.; Lednicer, D. Microbial degradation of (+)-camphor. J. Am. Chem. Soc. 1959, 81, 5507. [Google Scholar] [CrossRef]
- Gunsalus, I.C.; Conrad, H.E.; Trudgill, P.W.; Jacobson, L.A. Regulation of catabolic metabolism. Israel J. Med. Sci. 1965, 1, 1099–1119. [Google Scholar]
- Unger, B.P.; Gunsalus, I.C.; Sligar, S.G. Nucleotide sequence of the Pseudomonas putida cytrochrome P-450cam gene and its expression in Escherichia coli. J. Biol. Chem. 1986, 261, 1158–1163. [Google Scholar] [PubMed]
- Iwaki, H.; Grosse, S.; Bergeron, H.; Leisch, H.; Morley, K.; Hasegawa, Y.; Lau, P.C. Camphor pathway redox: functional recombinant expression of 2,5- and 3,6-diketocamphane monooxygenases in Pseudomonas putida ATCC 17453 with their cognate flavin reductase catalysing Baeyer-Villiger reactions. Appl. Environ. Microbiol. 2013, 79, 3282–3293. [Google Scholar] [CrossRef] [PubMed]
- Gunsalus, I.C.; Bertland, A.U.; Jacobson, L.A. Enzyme induction and repression in anabolic and catabolic pathways. Arch. Mikrobiol. 1967, 59, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Hartline, R.A.; Gunsalus, I.C. Induction specificity and catabolite repression of the early enzymes in camphor degradation by Pseudomonas putida. J. Bacteriol. 1971, 106, 468–478. [Google Scholar] [PubMed]
- Willetts, A. Structural studies and synthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol. 1997, 15, 55–62. [Google Scholar] [CrossRef]
- Ellis, H.R. The FMN-dependent two-component monooxygenase systems. Arch. Biochem. Biophys. 2010, 497, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rheinwald, J.G.; Chakrabarty, A.M.; Gunsalus, I.C. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc. Nat. Acad. Sci. USA 1973, 70, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Shaham, M.; Chakrabarty, A.M.; Gunsalus, I.C. Camphor plasmid-mediated chromosomal transfer in Pseudomonas putida. J. Bacteriol. 1973, 116, 944–949. [Google Scholar] [PubMed]
- Fieschi, F.; Niviere, V.; Frier, C.; Decout, J.-L.; Fontecave, M. The mechanism and substrate specificity of the NADPH: Flavin oxidoreductase from Escherichia coli. J. Biol. Chem. 1995, 270, 30392–30400. [Google Scholar] [CrossRef] [PubMed]
- Grogan, G. Microbial Biotransformations: Oxygenation of Cyclic Ketones by Baeyer-Villiger Monooxygenases from Pseudomonas putida NCIMB 10007. Ph. D. Thesis, University of Exeter, Exeter, UK, October 1995. [Google Scholar]
- Gagnon, R.; Grogan, G.; Roberts, S.M.; Villa, R.; Willetts, A. Enzymatic Baeyer-Villiger oxidation of some bicyclo[2.2.1]heptan-2-ones using monooxygenases from Pseudomonas putida NCIMB 10007: Enantioselective preparation of a precursor of azadirachtin. J. Chem. Soc. Perkin Trans. 1 1995, 12, 1505–1511. [Google Scholar] [CrossRef]
- Willetts, A.; Kelly, D.R. Multiple native flavin reductases in camphor-metabolising Pseudomonas putida NCIMB 10007: Functional interaction with two-component diketocamphane monooxygenase isoenzymes. Microbiology 2014, 160, 1784–1794. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Yeom, J.; Kang, Y.-S.; Kim, J.; Sung, J.-S.; Jeon, C.O.; Park, W. Molecular characterisation of FrpB (ferrodoxin-NADP:reductase) in Pseudomonas putida KT2440. J. Microbiol. Biotechnol. 2007, 17, 1504–1512. [Google Scholar] [PubMed]
- Yeom, J.; Jeon, C.O.; Madsen, E.L.; Park, W. Ferrodoxin-NADP (+) reductase from Pseudomonas putida functions as a ferric reductase. J. Bacteriol. 2009, 191, 1472–1479. [Google Scholar] [CrossRef] [PubMed]
- Poulos, T.P. Structural biology of heme monooxygenases. Biochem. Biophys. Res. Commun. 2005, 338, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Sevrioukova, I.F.; Poulos, T.P. Structural and mechanistic insights into the interaction of cytochrome P450A4 with bromoergocryptine, a type I ligand. J. Biol. Chem. 2011, 287, 3510–3517. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, Y.; Hass, M.A.; Kikui, Y.; Liu, W.M.; Olmez, B.; Skinner, S.P.; Blok, A.; Kloosterman, A.; Koteishi, H.; Lohr, F.; et al. The structure of the cytochrome P450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography. J. Mol. Biol. 2013, 425, 4353–4365. [Google Scholar] [CrossRef] [PubMed]
- Campbell, Z.T.; Baldwin, T.O. Fre is the major flavin reductase supporting bioluminescence from Vibrio harveyi luciferase in Escherichia coli. J. Biol. Chem. 2009, 284, 8322–8328. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Mortimer, M.W.; Fisher, T.S.; Kahn, M.J.; Brockma, F.J.; Xun, L. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH: Flavin mononucleotide oxidoreductase. J. Bacteriol. 1997, 179, 1112–1116. [Google Scholar] [PubMed]
- Matsubara, T.; Ohshiro, T.; Nishina, Y.; Izumi, Y. Purification, characterization, and overexpression of flavin reductase involved in dibenzothiophene desulfurization by Rhodococcus erythropolis D-1. Appl. Environ. Microbiol. 2001, 67, 1179–1184. [Google Scholar] [CrossRef] [PubMed]
- Thibaut, D.; Ratet, N.; Bisch, D.; Faucher, D.; Debussch, L.; Blanche, F. Purification of the two-enzyme system catalysing the oxidation of the D-proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J. Bacteriol. 1995, 177, 5199–5205. [Google Scholar] [PubMed]
- Parry, R.J.; Li, W. An NADPH: FAD oxidoreductase from the valinimycin producer, Streptomyces viridifaciens. J. Biol. Chem. 1997, 272, 23303–23311. [Google Scholar] [CrossRef] [PubMed]
- Valton, J.; Mathevon, C.; Fontecave, M.; Niviere, V.; Ballou, D.P. Mechanism and regulation of the two-component FMN-dependent monooxygenase ActVA-ActVB from Streptomyces coelicolor. J. Biol. Chem. 2008, 283, 10287–10296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beecher, J.E.; Willetts, A. Biotransformation of organic sulphides. Predictive active site models for sulfoxidation catalysed by the 2,5-diketocamphane 1,2-monooxygenase and 3,6-diketocamphane 1,6-monooxygenase, enantiocomplementary enzymes from Pseudomonas putida NCIMB 10007. Tetrahedron Asymm. 1998, 9, 1899–1916. [Google Scholar] [CrossRef]
- Lammerhofer, M.; Richter, M.; Wu, J.; Nogueira, R.; Bicker, W.; Lindner, W. Mixed-mode ion-exchangers and their comparative chromatograghic characterization in reverse-phase and hydrophilic interaction chromatography elution modes. J. Sep. Sci. 2008, 31, 2572–2588. [Google Scholar] [CrossRef] [PubMed]
- Gunsalus, I.C.; Wagner, G.C. Bacterial P-450cam methylene monooxygenase components: Cytochrome m, putidaredoxin and putidaredoxin reductase. Methods Enzymol. 1978, 52, 166–188. [Google Scholar] [PubMed]
- Halle, F.; Meyer, J.M. Iron release from ferrisiderophores. A multi-step mechanism involving a NADH/FMN oxidoreductase and a chemical reduction by FMNH2. Eur. J. Biochem. 1992, 209, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Filisetti, L.; Fontecave, M.; Niviere, V. Mechanism and substrate specificity of the flavin reductase ActVB from Streptomyces coelicolor. J. Biol. Chem. 2003, 278, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P.; Ballou, D.P.; Coon, M.J. Purified liver microsomal cytochrome P-450. Electron-accepting properties and oxidation-reduction potential. J. Biol. Chem. 1975, 250, 7405–7414. [Google Scholar] [PubMed]
- Massey, V.; Palmew, G. On the existence of spectrally distinct classes of flavoprotein semiquinones. A new method for the quantitative production of flavoprotein semiquinones. Biochemistry 1966, 5, 3181–3189. [Google Scholar] [CrossRef] [PubMed]
- Valton, J.; Filisetti, L.; Fontecave, M.; Niviere, V. The two-component flavin-dependent monooxygenase involved in actinorhodin biosynthesis in Streptomyces coelicolor. J. Biol. Chem. 2004, 279, 44362–44369. [Google Scholar] [CrossRef] [PubMed]
- Schröder, I.; Johnson, E.; de Vries, S. Microbial ferric iron reductases. FEMS Microbiol. Rev. 2003, 27, 427–447. [Google Scholar] [CrossRef]
- Yeom, J.; Park, W. Biochemical characterisation of ferrodoxin-NADP(+) reductase interaction with flavodoxin in Pseudomonas putida KT2440. BMB. Rep. 2012, 45, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Roome, P.W.; Philley, J.C.; Peterson, J.A. Purification and properties of putidaredoxin reductase. J. Biol. Chem. 1983, 258, 2593–2596. [Google Scholar] [PubMed]
- Vining, L.C. Roles of secondary metabolism in microbes. Ciba Found. Symp. 1992, 171, 184–194. [Google Scholar] [PubMed]
- Segel, I. Enzyme Kinetics; Wiley: New York, NY, USA, 1975. [Google Scholar]
- Reipa, V.G.; Holden, M.J.; Vilker, V.L. Association and redox properties of the putidaredoxin reductase-nicotinamide adenine dinucleotide complex. Biochemistry 2007, 46, 13235–13244. [Google Scholar] [CrossRef] [PubMed]
- Pardee, A.B.; Jacob, F.; Monod, J. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase in E. coli. J. Mol. Biol. 1959, 1, 165–187. [Google Scholar] [CrossRef]
- Fujita, M.; Aramaki, H.; Horiuchi, T.; Amemura, A. Transcription of the cam operon and camR genes in Pseudomonas putida PpG1. J. Bacteriol. 1993, 175, 6953–6958. [Google Scholar] [PubMed]
- Loescheke, A.; Thies, S. Pseudomonas putida—A versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 2015, 99, 6197–6214. [Google Scholar] [CrossRef] [PubMed]
- Gross, H.; Loper, J.E. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef] [PubMed]
- Conrad, H.E.; DuBus, R.; Namtvedt, M.J.; Gunsalus, I.C. Mixed function oxidation. II. Separation and properties of the enzymes catalysing camphor lactonization. J. Biol. Chem. 1965, 240, 495–503. [Google Scholar] [PubMed]
- Taylor, D.G.; Trudgill, P.W. Camphor revisited: Studies of 2,5-diketocamphane 1,2-monooxygenase from Pseudomonas putida ATCC 17453. J. Bacteriol. 1986, 165, 489–497. [Google Scholar] [PubMed]
- Katagari, M.; Ganguli, B.N.; Gunsalus, I.C. A soluble cytochrome P-450 functional in methylene hydroxylation. J. Biol. Chem. 1968, 243, 3543–3546. [Google Scholar]
- Beecher, J.E.; Grogan, G.; Roberts, S.; Willetts, A. Enantioselective oxidations by the diketocamphane monooxygenase isoenzymes from Pseudomonas putida. Biotechnol. Lett. 1996, 18, 571–576. [Google Scholar] [CrossRef]
- Ornston, L.N. Regulation of catabolic pathways in Pseudomonas. Bacteriol. Rev. 1971, 35, 87–116. [Google Scholar] [PubMed]
- Sucharitakul, J.; Phongsak, T.; Entsch, B.; Svasti, J.; Chaiyen, P.; Ballou, D.P. Kinetics of a two-component p-hydroxyphenylacetate hydroxylase explains how reduced flavin is transferred from the reductase to the oxygenase. Biochemistry 2007, 46, 8611–8623. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.J.; Yoon, H.J.; Lee, B.; Sub, S.W. Crystal structure of chorismate synthase: A novel FMN-binding protein fold and functional insights. J. Mol. Biol. 2004, 336, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, D.E.; Binda, C.; Mattevi, A. The FAD-binding sites of human monoamine oxidases A and B. Neuro. Toxicol. 2004, 25, 63–72. [Google Scholar] [CrossRef]
- Chaiyen, P.; Fraaije, M.W.; Mattevi, A. The enigmatic reaction of flavins with oxygen. Trends Biochem. Sci. 2012, 37, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Thotsaporn, K.; Chenprakhon, P.; Sucharitakul, J.; Mattevi, A.; Chaiyen, P. Stabilization of C4a–hydroperoxyflavin in two-component flavin-dependent monooxygenase is achieved through interactions at flavin N5 and C4a atoms. J. Biol. Chem. 2011, 286, 28170–28180. [Google Scholar] [CrossRef] [PubMed]
- Isupov, M.N.; Schroder, E.; Gibson, R.P.; Beecher, J.; Donadio, G.; Saneei, V.; Dcunha, S.A.; McGhie, E.J.; Sayer, C.; Davenport, C.F.; et al. The oxygenating constituent of 3,6-diketocamphane monooxygenase from the CAM plasmid of Pseudomonas putida: The first crystal structure of a type II Baeyer-Villiger monooxygenase. Acta. Cryst. D. 2015, 71, 2344–2351. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.J.; Thompson, T.B.; Thoden, J.B.; Baldwin, T.O.; Rayment, I. The 1.5 A resolution crystal structure of bacterial luciferase in low salt conditions. J. Biol. Chem. 1996, 271, 21956–21968. [Google Scholar] [PubMed]
- Waddle, J.; Baldwin, T.O. Individual α and β subunits of bacterial luciferase exhibit bioluminescence activity. Biochem. Biophys. Res. Commun. 1991, 178, 1188–1193. [Google Scholar] [CrossRef]
- Campbell, Z.T.; Weichsel, S.; Montfort, W.R.; Baldwin, T.D. Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the β subunit. Biochemistry 2009, 48, 6085–6094. [Google Scholar] [CrossRef] [PubMed]
- Aufhammer, S.W.; Warkentin, E.; Berk, H.; Shima, S.; Thauer, R.K.; Ermier, U. Coenzyme binding in F420-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family. Structure 2004, 12, 361–370. [Google Scholar] [CrossRef] [PubMed]

| Enzyme Combination | (+)Camphor % Oxidation (%ee Lactone Product) 180 m 360 m | (−)Camphor % Oxidation (%ee Lactone Product) 180 m 360 m | 2,5-DKC % Oxidation (%ee Lactone Product) 180 m 360 m | 3,6-DKC % Oxidation (%ee Lactone Product) 180 m 360 m | (+)Fenchone % Oxidation (%ee Lactone Product) 180 m 360 m | (−)Fenchone % Oxidation (%ee Lactone Product) 180 m 360 m |
|---|---|---|---|---|---|---|
| 25DKCMO + | 58 90 | 0 0 | 66 100 | 0 0 | 0 0 | 0 0 |
| Fred | (96) (98) | (na) (na) | ||||
| 25DKCMO + | 62 95 | 0 0 | 69 100 | 0 0 | 0 0 | 0 0 |
| Frp1 & 2 | (94) (98) | (na) (na) | ||||
| 25DKCMO + | 48 86 | 0 0 | 54 90 | 0 0 | 0 0 | 0 0 |
| PdR | (92) (97) | (na) (na) | ||||
| 36DKCMO + | 0 0 | 60 92 | 0 0 | 76 100 | 0 0 | 0 0 |
| Fred | (94) (96) | (na) (na) | ||||
| 36DKCMO + | 0 0 | 58 95 | 0 0 | 76 100 | 0 0 | 0 0 |
| Frp1 & 2 | (96) (98) | (na) (na) | ||||
| 36DKCMO + | 0 0 | 49 86 | 0 0 | 62 93 | 0 0 | 0 0 |
| PdR | (93) (97) | (na) (na) |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willetts, A.; Kelly, D. Flavin-Dependent Redox Transfers by the Two-Component Diketocamphane Monooxygenases of Camphor-Grown Pseudomonas putida NCIMB 10007. Microorganisms 2016, 4, 38. https://doi.org/10.3390/microorganisms4040038
Willetts A, Kelly D. Flavin-Dependent Redox Transfers by the Two-Component Diketocamphane Monooxygenases of Camphor-Grown Pseudomonas putida NCIMB 10007. Microorganisms. 2016; 4(4):38. https://doi.org/10.3390/microorganisms4040038
Chicago/Turabian StyleWilletts, Andrew, and David Kelly. 2016. "Flavin-Dependent Redox Transfers by the Two-Component Diketocamphane Monooxygenases of Camphor-Grown Pseudomonas putida NCIMB 10007" Microorganisms 4, no. 4: 38. https://doi.org/10.3390/microorganisms4040038






