Modeling Lactic Fermentation of Gowé Using Lactobacillus Starter Culture
Abstract
:1. Introduction
2. Materials and Methods
2.1. Raw Material
2.2. Microbial Strains
2.3. Microbiological Methods
2.3.1. Batch Cultures
2.3.2. Plate Counting Methods
2.4. Chemical Analyses
2.5. Mathematical Modeling
2.5.1. Primary and Secondary Models
2.5.2. Modeling Lactic Acid Production
2.5.3. Modeling the pH Value of Gowé
2.6. Statistical Methods
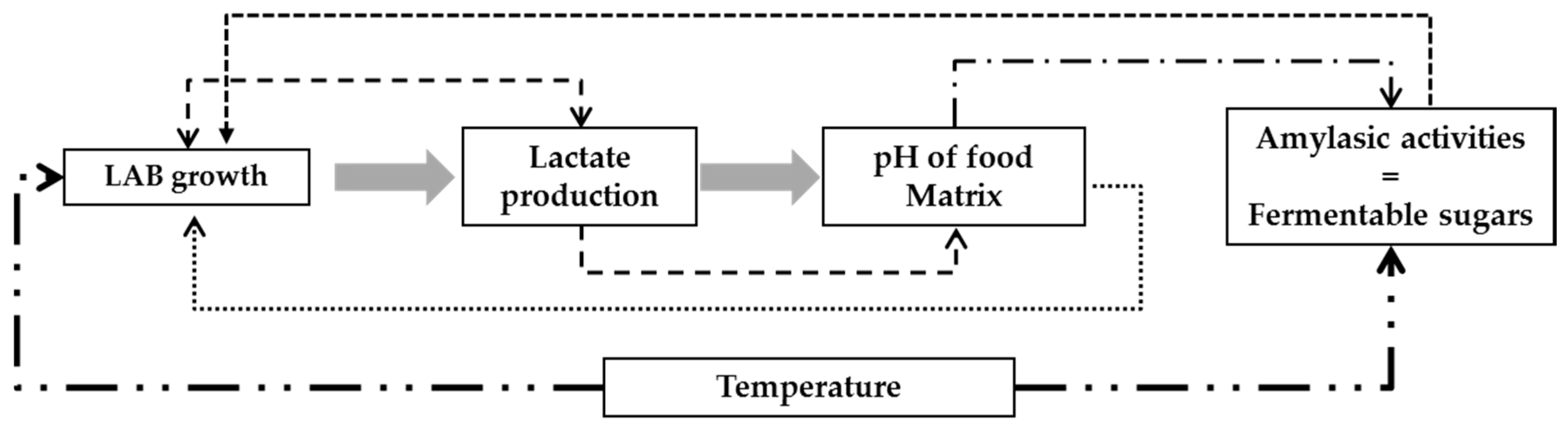
3. Results and Discussion
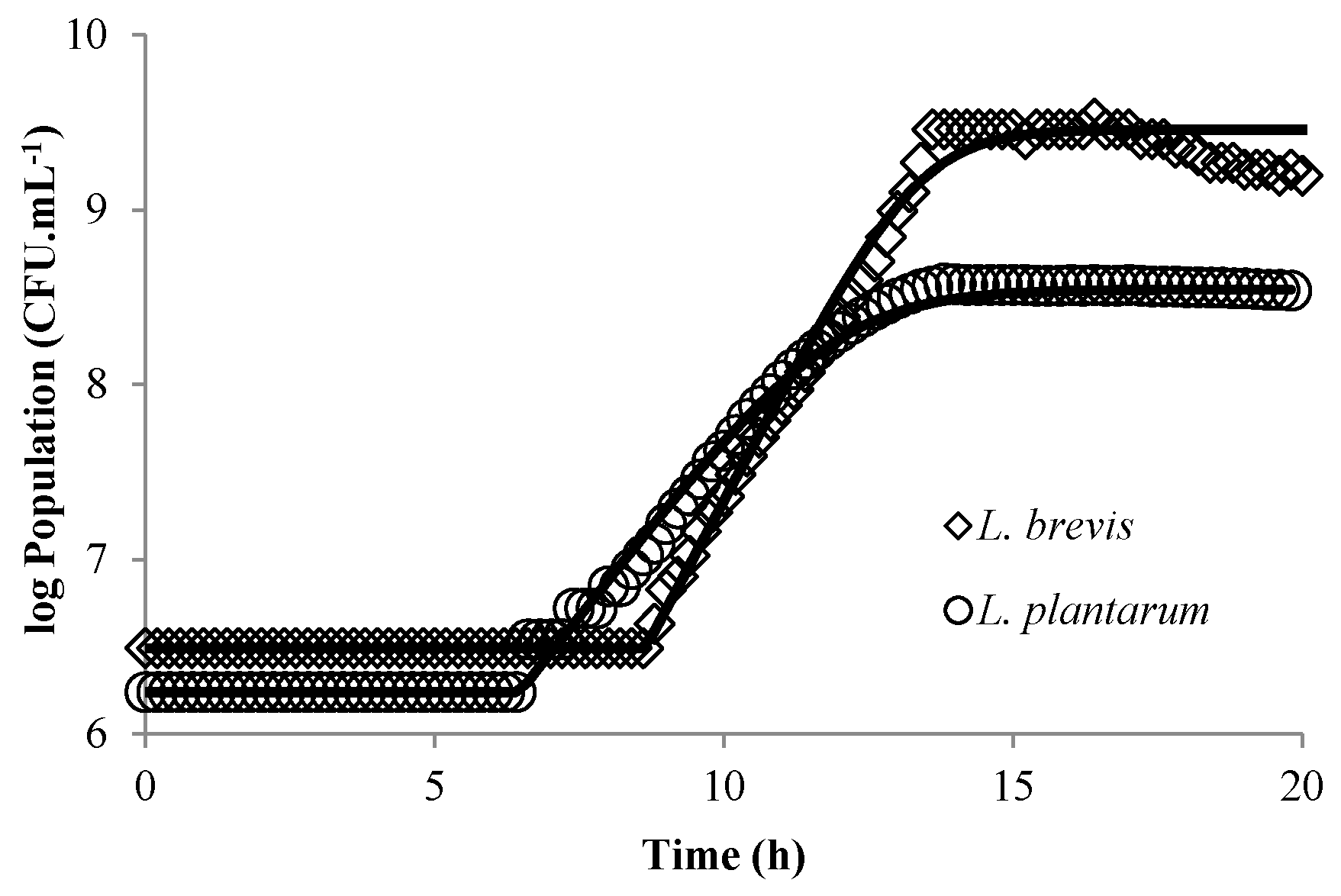
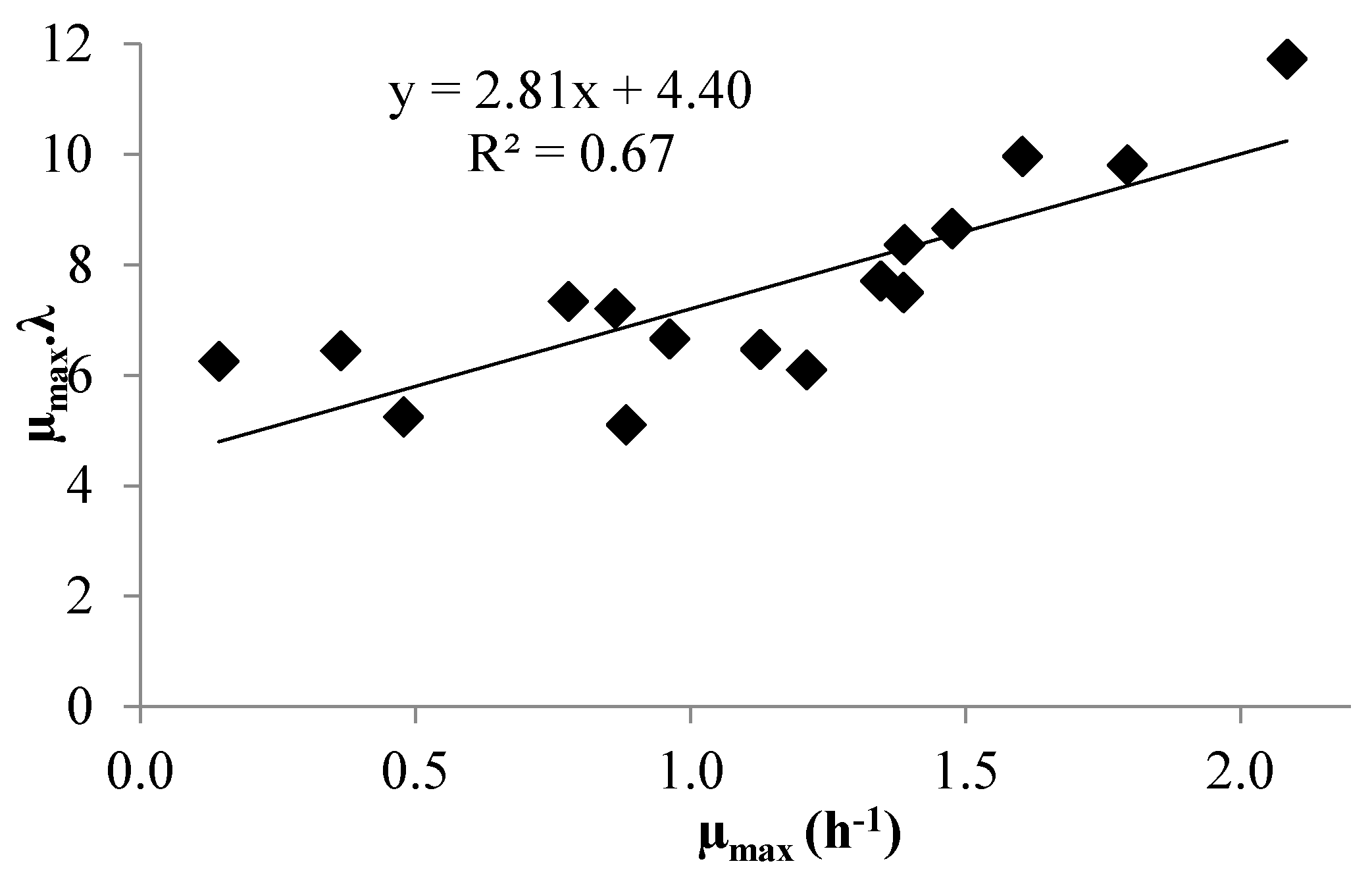
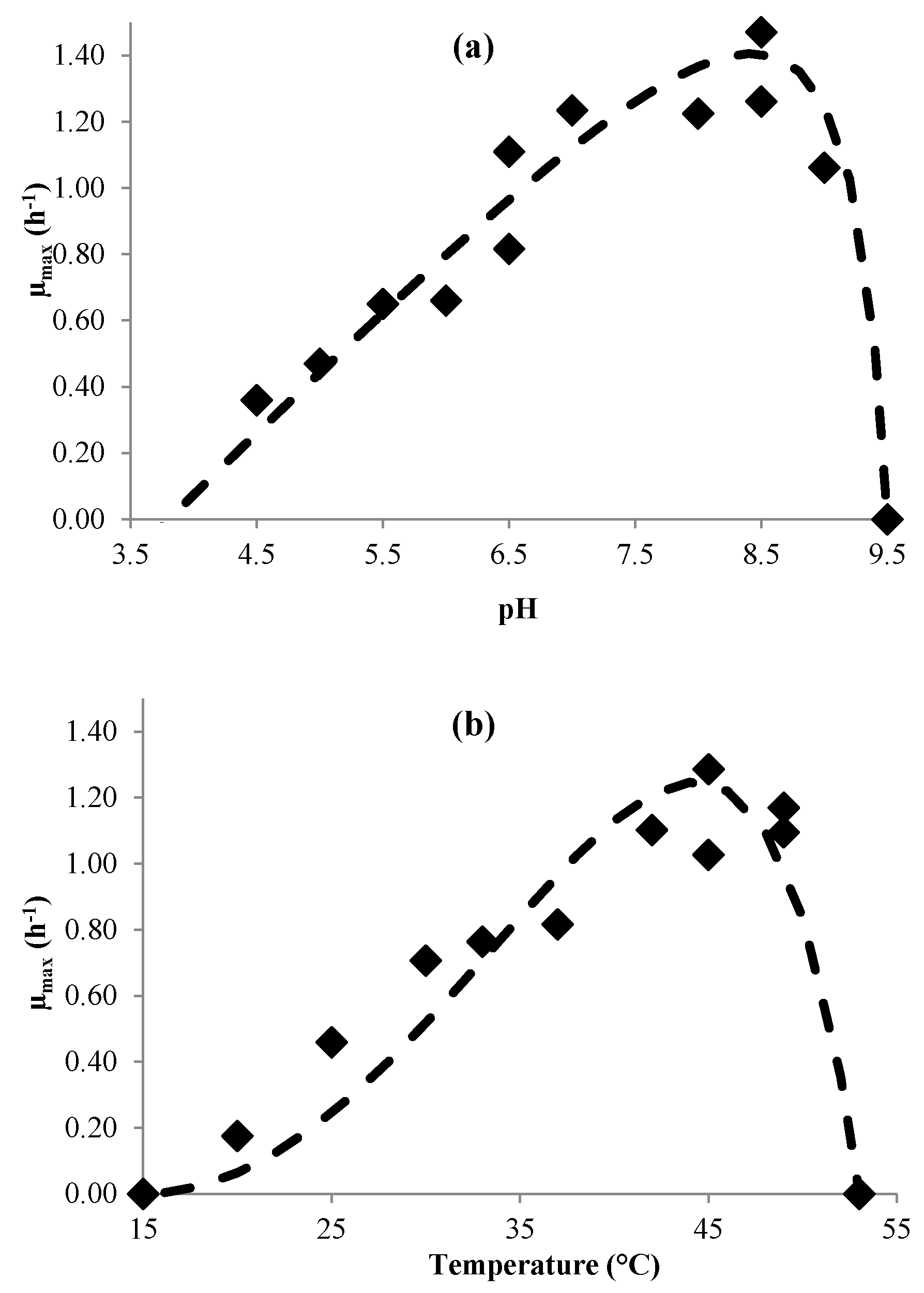
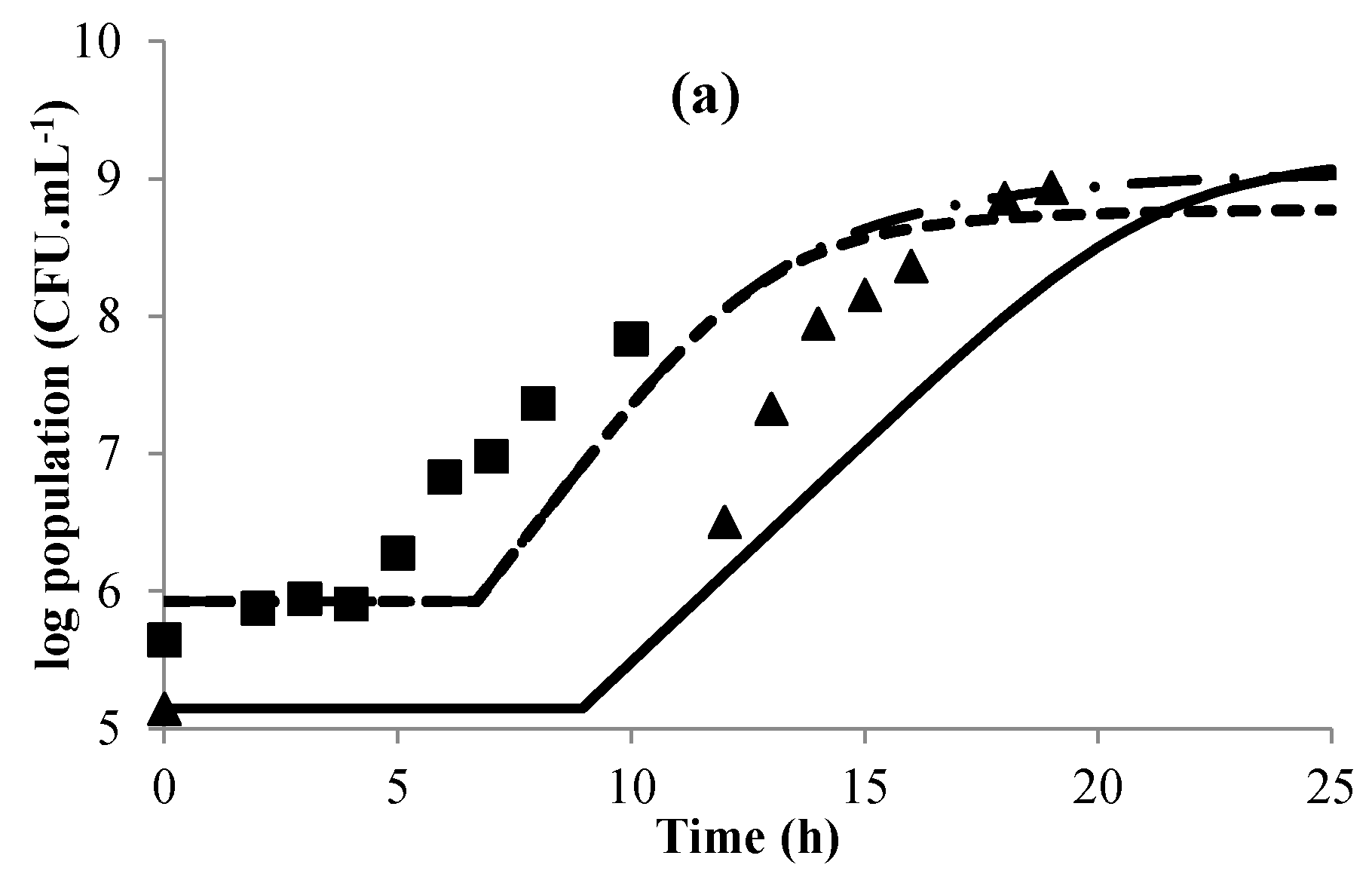
3.1. Modeling Bacterial Growth
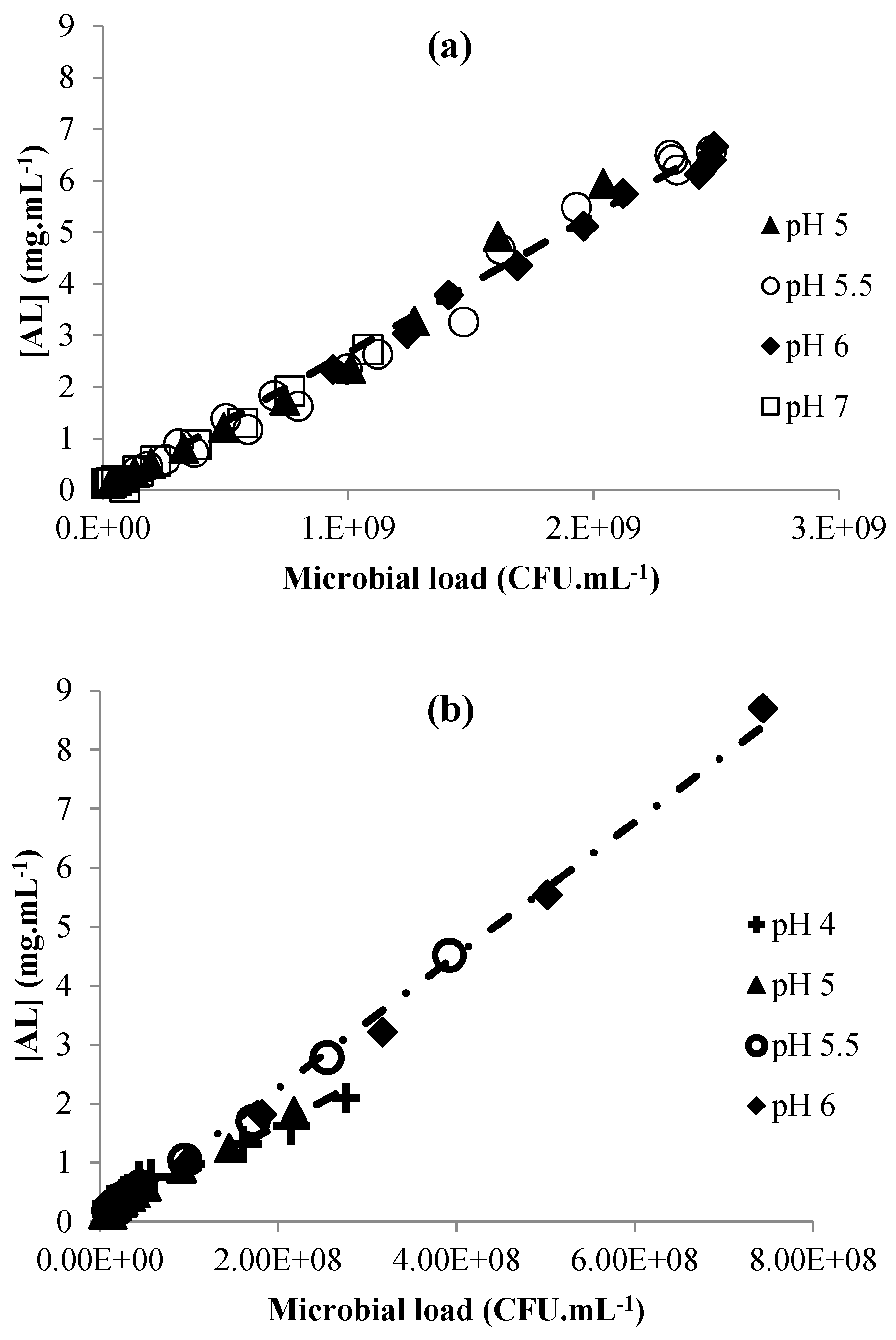
3.2. Acid Lactic Production
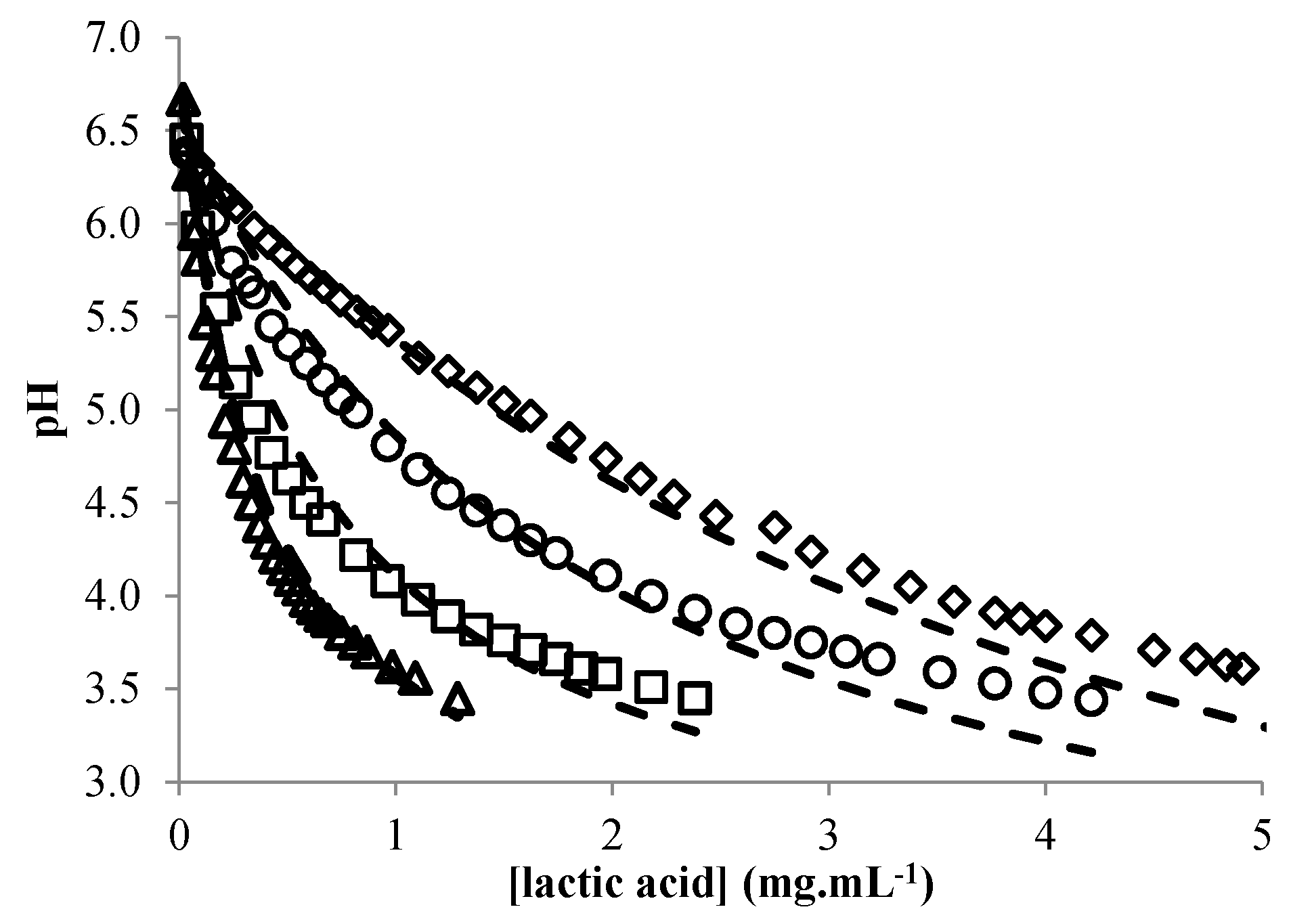
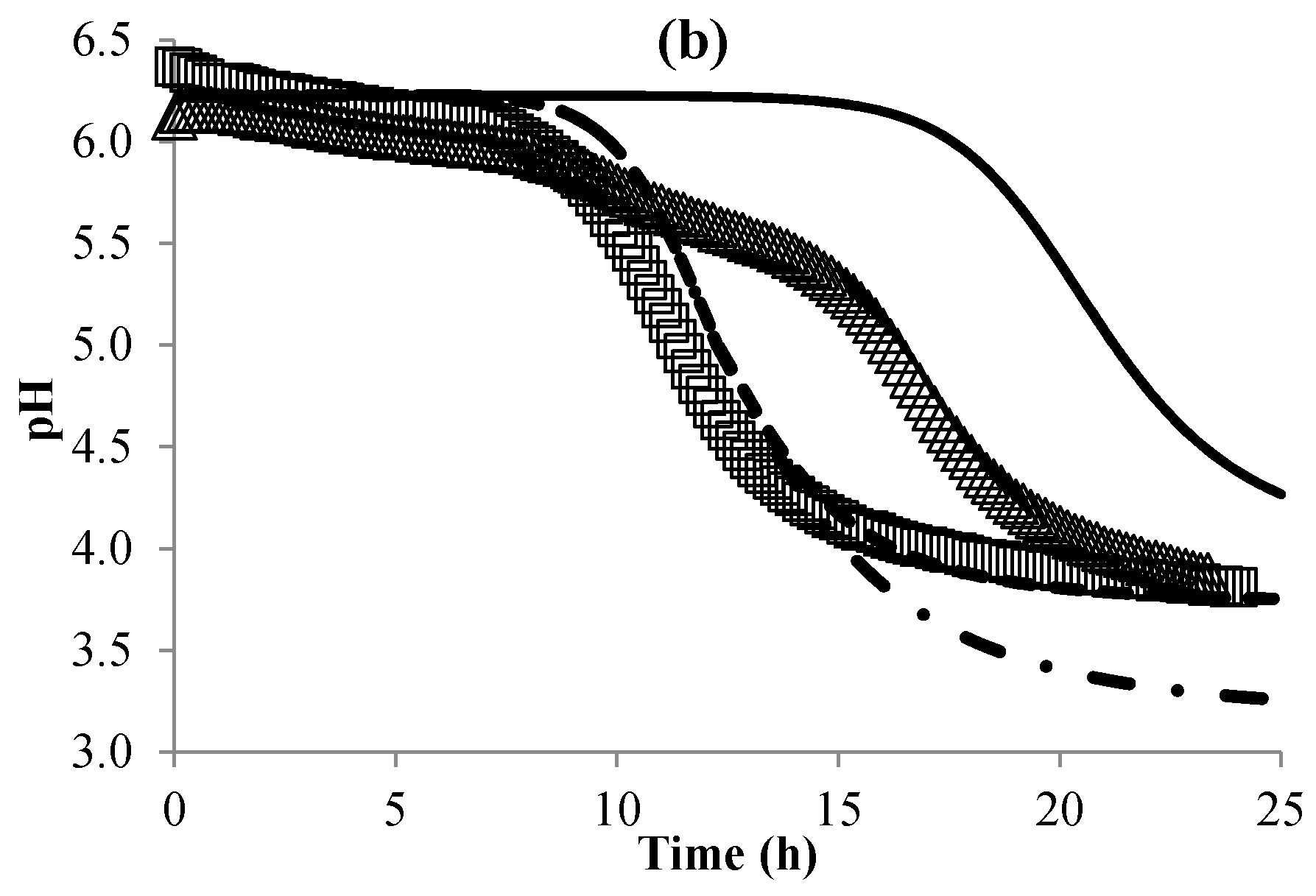
3.3. Modeling the pH of Raw Material
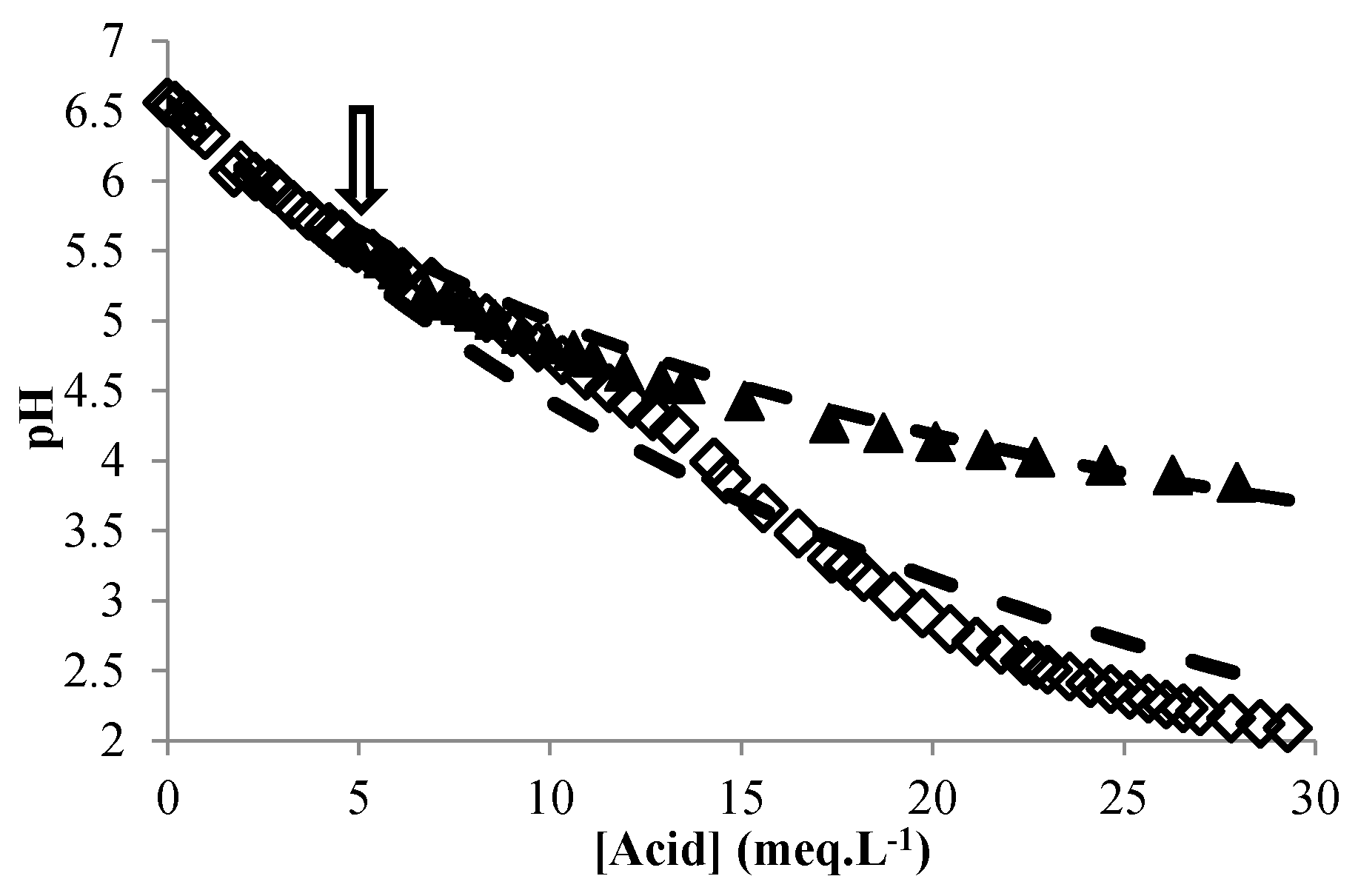
3.4. Validation of the Global Model
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cocolin, L.; Ercolini, D. Molecular Techniques in the Microbial Ecology of Fermented Foods; Springer Science & Business Media: New York, NY, USA, 2008. [Google Scholar]
- Aka, S.; Konan, G.; Fokou, G.; Dje, K.M.; Bonfoh, B. Review on African traditional cereal beverages. Am. J. Res. Commun. 2014, 2, 104–153. [Google Scholar]
- Guyot, J.-P. Cereal-based fermented foods in developing countries: Ancient foods for modern research. Int. J. Food Sci. Technol. 2012, 47, 1109–1114. [Google Scholar] [CrossRef]
- Achi, O.K. The potential for upgrading traditional fermented foods through biotechnology. Afr. J. Biotechnol. 2013, 4, 375–380. [Google Scholar]
- Holzapfel, W.H. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 2002, 75, 197–212. [Google Scholar] [CrossRef]
- Caplice, E.; Fitzgerald, G.F. Food fermentations: Role of microorganisms in food production and preservation. Int. J. Food Microbiol. 1999, 50, 131–149. [Google Scholar] [CrossRef]
- Leroy, F.; Degeest, B.; de Vuyst, L. A novel area of predictive modelling: Describing the functionality of beneficial microorganisms in foods. Int. J. Food Microbiol. 2002, 73, 251–259. [Google Scholar] [CrossRef]
- Kristo, E.; Biliaderis, C.G.; Tzanetakis, N. Modelling of the acidification process and rheological properties of milk fermented with a yogurt starter culture using response surface methodology. Food Chem. 2003, 83, 437–446. [Google Scholar] [CrossRef]
- Bonatsou, S.; Benítez, A.; Rodríguez-Gómez, F.; Panagou, E.Z.; Arroyo-López, F.N. Selection of yeasts with multifunctional features for application as starters in natural black table olive processing. Food Microbiol. 2015, 46, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Rosso, L.; Lobry, J.R.; Flandrois, J.P. An unexpected correlation between cardinal temperatures of microbial growth highlighted by a new model. J. Theor. Biol. 1993, 162, 447–463. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, J.; Prats, C.; López, D.; Vives-Rego, J. Mathematical modelling methodologies in predictive food microbiology: A SWOT analysis. Int. J. Food Microbiol. 2009, 134, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Ratkowsky, D.A.; Lowry, R.K.; Mc Meekin, T.A.; Stokes, A.N.; Chandler, R.E. Model for bacterial culture growth rate throughout the entire biokinetic temperature range. J. Bacteriol. 1983, 154, 1222–1226. [Google Scholar] [PubMed]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Rosso, L. Modélisation et Microbiologie Prévisionnelle: Élaboration d’un Nouvel Outil Pour l’Agro-Alimentaire; Université Claude Bernard-Lyon I: Lyon, France, 1995. [Google Scholar]
- Lebert, I.; Robles-Olvera, A.; Lebert, A. Application of polynomial models to predict growth of mixed cultures of Pseudomonas spp. and Listeria in meat. Int. J. Food Microbiol. 2000, 61, 27–39. [Google Scholar] [CrossRef]
- Watier, D.; Dubourguier, H.C.; Leguerinel, I.; Hornez, J.P. Response surface models to describe the effects of temperature, pH, and ethanol concentration on growth kinetics and fermentation end products of a Pectinatus. sp. Appl. Environ. Microbiol. 1996, 62, 1233–1237. [Google Scholar] [PubMed]
- Zwietering, M.H.; Wijtzes, T.; Rombouts, F.M.; van’t Riet, K. A decision support system for prediction of microbial spoilage in foods. J. Ind. Microbiol. 1993, 12, 324–329. [Google Scholar] [CrossRef]
- Le Marc, Y.; Huchet, V.; Bourgeois, C.M.; Guyonnet, J.P.; Mafart, P.; Thuault, D. Modelling the growth kinetics of Listeria as a function of temperature, pH and organic acid concentration. Int. J. Food Microbiol. 2002, 73, 219–237. [Google Scholar] [CrossRef]
- Degeest, B.; de Vuyst, L. Indication that the nitrogen source influences both amount and size of exopolysaccharides produced by Streptococcus thermophilus LY03 and modelling of the bacterial growth and exopolysaccharide production in a complex medium. Appl. Environ. Microbiol. 1999, 65, 2863–2870. [Google Scholar] [PubMed]
- Vereecken, K.M.; van Impe, J.F. Analysis and practical implementation of a model for combined growth and metabolite production of lactic acid bacteria. Int. J. Food Microbiol. 2002, 73, 239–250. [Google Scholar] [CrossRef]
- Michodjèhoun-Mestres, L.; Joseph, H.; Joseph, D.; Christian, M. Physical, chemical and microbiological changes during natural fermentation of “gowé”, a sprouted or non-sprouted sorghum beverage from West-Africa. Afr. J. Biotechnol. 2005, 4, 487–496. [Google Scholar]
- Vieira-Dalodé, G.; Madodé, Y.E.; Hounhouigan, J.; Jespersen, L.; Jakobsen, M. Use of starter cultures of lactic acid bacteria and yeasts as inoculum enrichment for the production of gowé, a sour beverage from Benin. Afr. J. Microbiol. Res. 2008, 2, 179–186. [Google Scholar]
- Adinsi, L.; Vieira-Dalode, G.; Akissoe, N.; Anihouvi, V.; Mestres, C.; Jacobs, A.; Dlamini, N.; Pallet, D.; Hounhouigan, D.J. Processing and quality attributes of gowé: A malted and fermented cereal-based beverage from Benin. Food Chain. 2014, 4, 171–183. [Google Scholar]
- Vieira-Dalodé, G.; Jespersen, L.; Hounhouigan, J.; Moller, P.L.; Nago, C.M.; Jakobsen, M. Lactic acid bacteria and yeasts associated with gowé production from sorghum in Bénin. J. Appl. Microbiol. 2007, 103, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Greppi, A.; Rantisou, K.; Padonou, W.; Hounhouigan, J.; Jespersen, L.; Jakobsen, M.; Cocolin, L. Yeast dynamics during spontaneous fermentation of mawè and tchoukoutou, two traditional products from Benin. Int. J. Food Microbiol. 2013, 165, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Adinsi, L.; Akissoé, N.H.; Dalodé-Vieira, G.; Anihouvi, V.B.; Fliedel, G.; Mestres, C.; Hounhouigan, J.D. Sensory evaluation and consumer acceptability of a beverage made from malted and fermented cereal: Case of gowé from Benin. Food Sci. Nutr. 2015, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mestres, C.; Nguyen, T.C.; Adinsi, L.; Hounhouigan, J.; Fliedel, G.; Loiseau, G. The interaction between starch hydrolysis and acidification kinetic determines the quality of a malted and fermented sorghum beverage. J. Cereal Sci. 2015, 63, 8–13. [Google Scholar] [CrossRef]
- Vieira-Dalodé, G.; Akissoé, N.; Hounhouigan, D.J.; Jakobsen, M.; Mestres, C. Dextrin, sugar and organic acid profiles of spontaneous and modified gowé: A malted and fermented sorghum beverage from Benin. Int. J. Food Sci. Technol. 2015, 50, 549–557. [Google Scholar] [CrossRef]
- Van Impe, J.; Poschet, F.; Geeraerd, A.; Vereecken, K. Towards a novel class of predictive microbial growth models. Int. J. Food Microbiol. 2005, 100, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Rosso, L.; Lobry, J.R.; Bajard, S.; Flandrois, J.P. Convenient model to describe the combined effects of temperature and pH on microbial growth. Appl. Environ. Microbiol. 1995, 61, 610–616. [Google Scholar] [PubMed]
- Coroller, L.; Guerrot, V.; Huchet, V.; Lemarc, Y.; Mafart, P.; Sohier, D.; Thuault, D. Modelling the influence of single acid and mixture on bacterial growth. Int. J. Food Microbiol. 2005, 100, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Gätje, G.; Gottschalk, G. Limitation of growth and lactic acid production in batch and continuous cultures of Lactobacillus helveticus. Appl. Microbiol. Biotechnol. 1991, 34, 446–449. [Google Scholar] [CrossRef]
- Swinnen, I.; Bernaerts, K.; Dens, E.J.J.; Geeraerd, A.H.; van Impe, J.F. Predictive modelling of the microbial lag phase: A review. Int. J. Food Microbiol. 2004, 94, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Charalampopoulos, D.; Vázquez, J.A.; Pandiella, S.S. Modelling and validation of Lactobacillus plantarum fermentations in cereal-based media with different sugar concentrations and buffering capacities. Biochem. Eng. J. 2009, 44, 96–105. [Google Scholar] [CrossRef] [Green Version]
- Augustin, J.-C.; Brouillaud-Delattre, A.; Rosso, L.; Carlier, V. Significance of Inoculum size in the lag time of Listeria monocytogenes. Appl. Environ. Microbiol. 2000, 66, 1706–1710. [Google Scholar] [CrossRef] [PubMed]
- Wijtzes, T.; de Wit, J.C.; Van’t, R.; Zwietering, M.H. Modelling bacterial growth of Lactobacillus curvatus as a function of acidity and temperature. Appl. Environ. Microbiol. 1995, 61, 2533–2539. [Google Scholar] [PubMed]
- Liu, B.; Yang, M.; Qi, B.; Chen, X.; Su, Z.; Wan, Y. Optimizing l-(+)-lactic acid production by thermophile Lactobacillus plantarum As.1.3 using alternative nitrogen sources with response surface method. Biochem. Eng. J. 2010, 52, 212–219. [Google Scholar] [CrossRef]
- Sheeladevi, A. Lactic acid production using lactic acid bacteria under optimized conditions. Int. J. Pharm. Biol. Arch. 2011, 2, 6. [Google Scholar]
- Ogunbanwo, S.T.; Sanni, A.I.; Onilude, A.A. Influence of cultural conditions on the production of bacteriocin by Lactobacillus brevis OG1. Afr. J. Biotechnol. 2003, 2, 179–184. [Google Scholar]
- Mihhalevski, A.; Sarand, I.; Viiard, E.; Salumets, A.; Paalme, T. Growth characterization of individual rye sourdough bacteria by isothermal microcalorimetry. J. Appl. Microbiol. 2011, 110, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Lelong, B.; Raimbault, M. Influence of pH and initial lactate concentration on the growth of Lactobacillus plantarum. Appl. Microbiol. Biotechnol. 1991, 36, 96–99. [Google Scholar] [CrossRef]
- Lambert, R.J.W. A new model for the effect of pH on microbial growth: An extension of the Gamma hypothesis. J. Appl. Microbiol. 2011, 110, 61–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogunbanwo, S.T.; Okanlawon, B.M. Influence of nutrients utilization and cultivation conditions on the production of lactic acid by homolactic fermenters. Biotechnology 2009, 8, 107–113. [Google Scholar] [CrossRef]
- Passos, F.V.; Fleming, H.P.; Ollis, D.F.; Hassan, H.M.; Felder, R.M. Modeling the specific growth rate of Lactobacillus plantarum in cucumber extract. Appl. Microbiol. Biotechnol. 1993, 40, 143–150. [Google Scholar] [CrossRef]
- Pelinescu, D.-R.; Sasarman, E.; Chifiriuc, M.-C.; Stoica, I.; Nohit, A.-M.; Avram, I.; Serbancea, F.; Dimov, T.V. Isolation and identification of some Lactobacillus and Enterococcus strains by a polyphasic taxonomical approach. Romanian Biotechnol. Lett. 2009, 14, 4225–4233. [Google Scholar]
- Zhou, K.; Zeng, Y.; Han, X.; Liu, S. Modelling growth and bacteriocin production by Lactobacillus plantarum BC-25 in response to temperature and pH in batch fermentation. Appl. Biochem. Biotechnol. 2015, 176, 1627–1637. [Google Scholar] [CrossRef] [PubMed]
- Dubernet, S.; Desmasures, N.; Guéguen, M. A PCR-based method for identification of lactobacilli at the genus level. FEMS Microbiol. Lett. 2002, 214, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Ferrada, B.M.; Hollmann, A.; Delfederico, L.; Valdés La Hens, D.; Caballero, A.; Semorile, L. Patagonian red wines: Selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J. Microbiol. Biotechnol. 2013, 29, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, G. Bacterial Metabolism, 2nd ed.; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Thompson, J.; Gentry-Weeks, C.R. Métabolisme des sucres par les bactéries lactiques. In Bacteries lactiques: Aspects Fondamentaux et Technologiques; de Roissart, H., Luquet, F.M., Eds.; Lorica: Uriage, France, 1994. [Google Scholar]
- Fu, W.; Mathews, A.P. Lactic acid production from lactose by Lactobacillus plantarum: Kinetic model and effects of pH, substrate, and oxygen. Biochem. Eng. J. 1999, 3, 163–170. [Google Scholar] [CrossRef]
- Wilson, P.D.G.; Wilson, D.R.; Waspe, C.R. Weak acids: Dissociation in complex buffering systems and partitioning into oils. J. Sci. Food Agric. 2000, 80, 471–476. [Google Scholar] [CrossRef]
- Gonçalves, L.M.D.; Ramos, A.; Almeida, J.S.; Xavier, A.M.R.B.; Carrondo, M.J.T. Elucidation of the mechanism of lactic acid growth inhibition and production in batch cultures of Lactobacillus rhamnosus. Appl. Microbiol. Biotechnol. 1997, 48, 346–350. [Google Scholar]
- Gadgil, C.J.; Venkatesh, K.V. Structured Model for Batch Culture Growth of Lactobacillus bulgaricus. J. Chem. Technol. Biotechnol. 1997, 69, 89–93. [Google Scholar] [CrossRef]
| Modeled Variable | Equation | Parameter | Value ± CI (L. brevis) | Value ± CI (L. plantarum) |
|---|---|---|---|---|
| µmax | (3) | pHmin pHmax pHopt µopt (h−1) Tmin (°C) Tmax (°C) Topt (°C) | 3.8 9.3 8.3 (±0.3) 1.8 (±0.2) 15 53 44.6 (±1.2) | 3.2 9.2 8.3 (±0.3) 1.4 (±0.1) 12 52 37.1 (±3.2) |
| (4) | µmax ratio | 1.57 (±0.1) | 1.36 (±0.06) | |
| MIC | (5) | [AH] mM | 23.73 | 35.59 |
| λ | (6) | α | 4.4 (±1.2) | 1.17 (±1.4) |
| β | 2.8 (±1.0) | 5.6 (±1.5) | ||
| Lactic acid production rate | (7) | Y(lactic)/N (10−9 mg·CFU−1) | 2.71 (±0.08) | 11.3 (±0.3) * 8.5 (±0.6) $ |
| pHRM | (10) | φ | 6.30 (±0.006) | |
| ε | 0.028 (±0003) | |||
| BP | (11) | (mL·g−1) | 0.16 (±0.02) | |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De J. C. Munanga, B.; Loiseau, G.; Grabulos, J.; Mestres, C. Modeling Lactic Fermentation of Gowé Using Lactobacillus Starter Culture. Microorganisms 2016, 4, 44. https://doi.org/10.3390/microorganisms4040044
De J. C. Munanga B, Loiseau G, Grabulos J, Mestres C. Modeling Lactic Fermentation of Gowé Using Lactobacillus Starter Culture. Microorganisms. 2016; 4(4):44. https://doi.org/10.3390/microorganisms4040044
Chicago/Turabian StyleDe J. C. Munanga, Bettencourt, Gérard Loiseau, Joël Grabulos, and Christian Mestres. 2016. "Modeling Lactic Fermentation of Gowé Using Lactobacillus Starter Culture" Microorganisms 4, no. 4: 44. https://doi.org/10.3390/microorganisms4040044





