Effects of Dietary Yogurt on the Healthy Human Gastrointestinal (GI) Microbiome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enumeration and Confirmation of Probiotic Bacteria in Yogurt
2.2. Experimental Design and Sampling Collection
2.3. T-RFLP Analyses and Sanger Sequencing
2.4. Next Generation Sequencing
3. Results
3.1. Viable Probiotic Bacteria in Dietary Yogurt
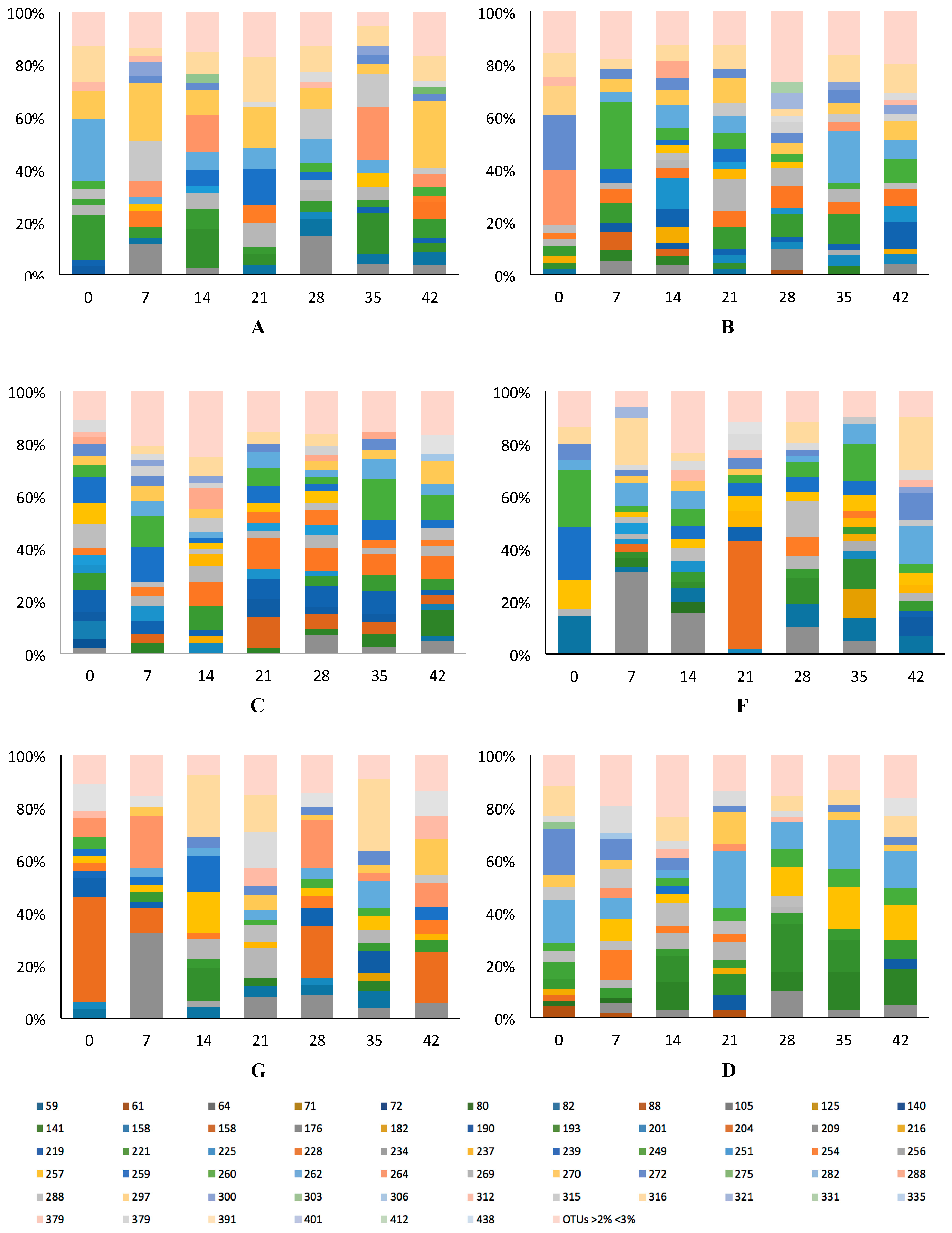
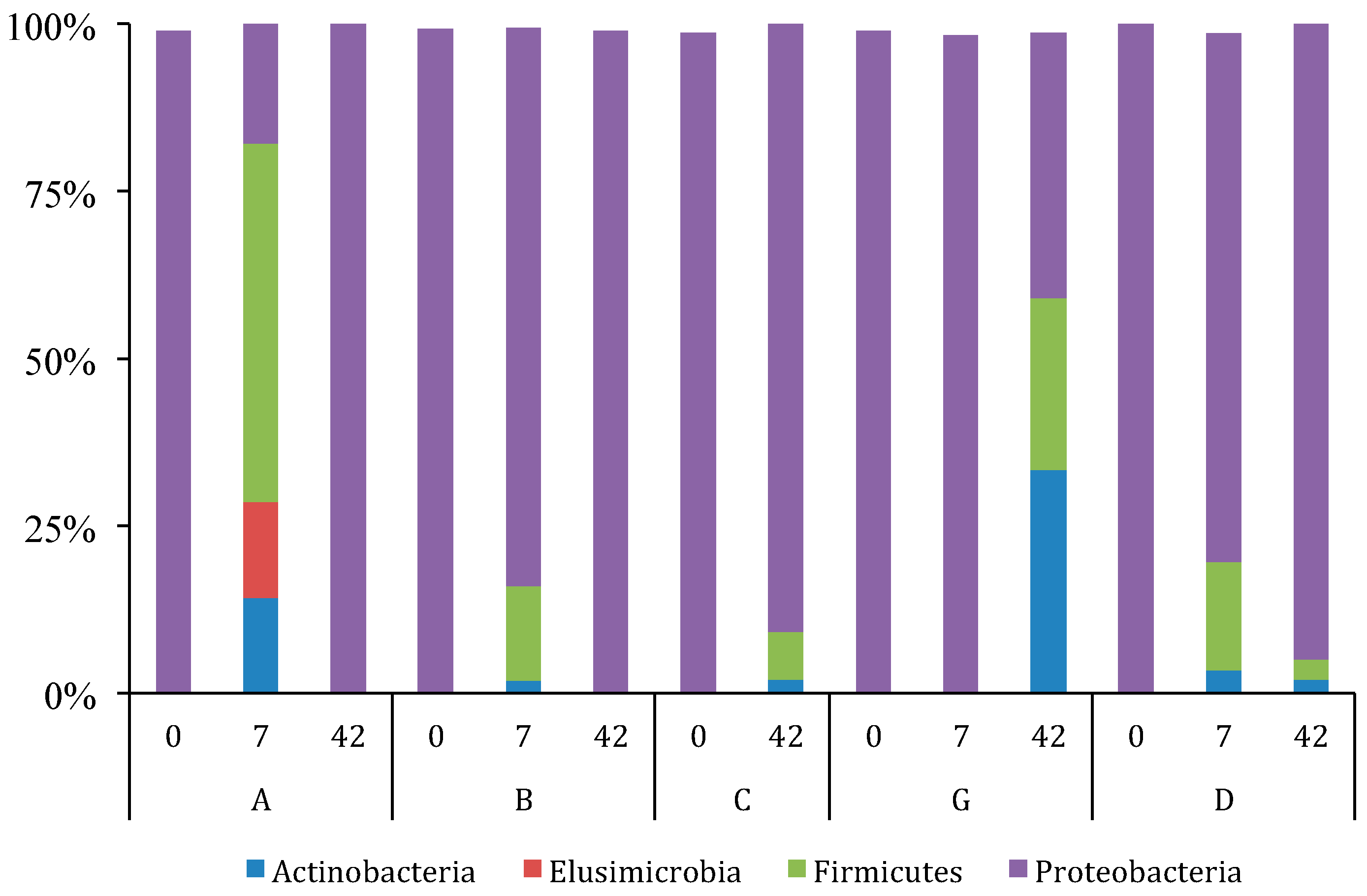
3.2. GI Bacterial Communities
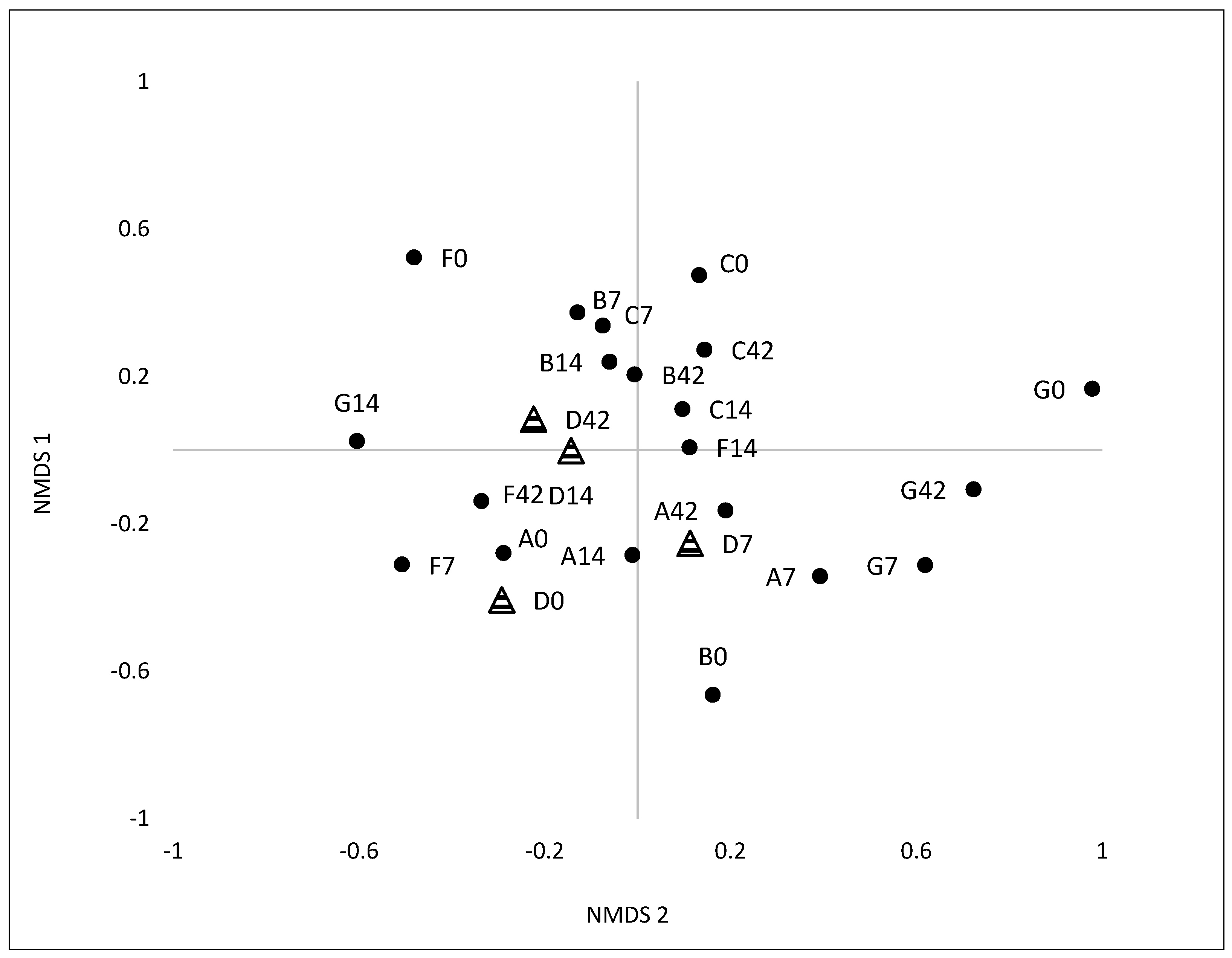
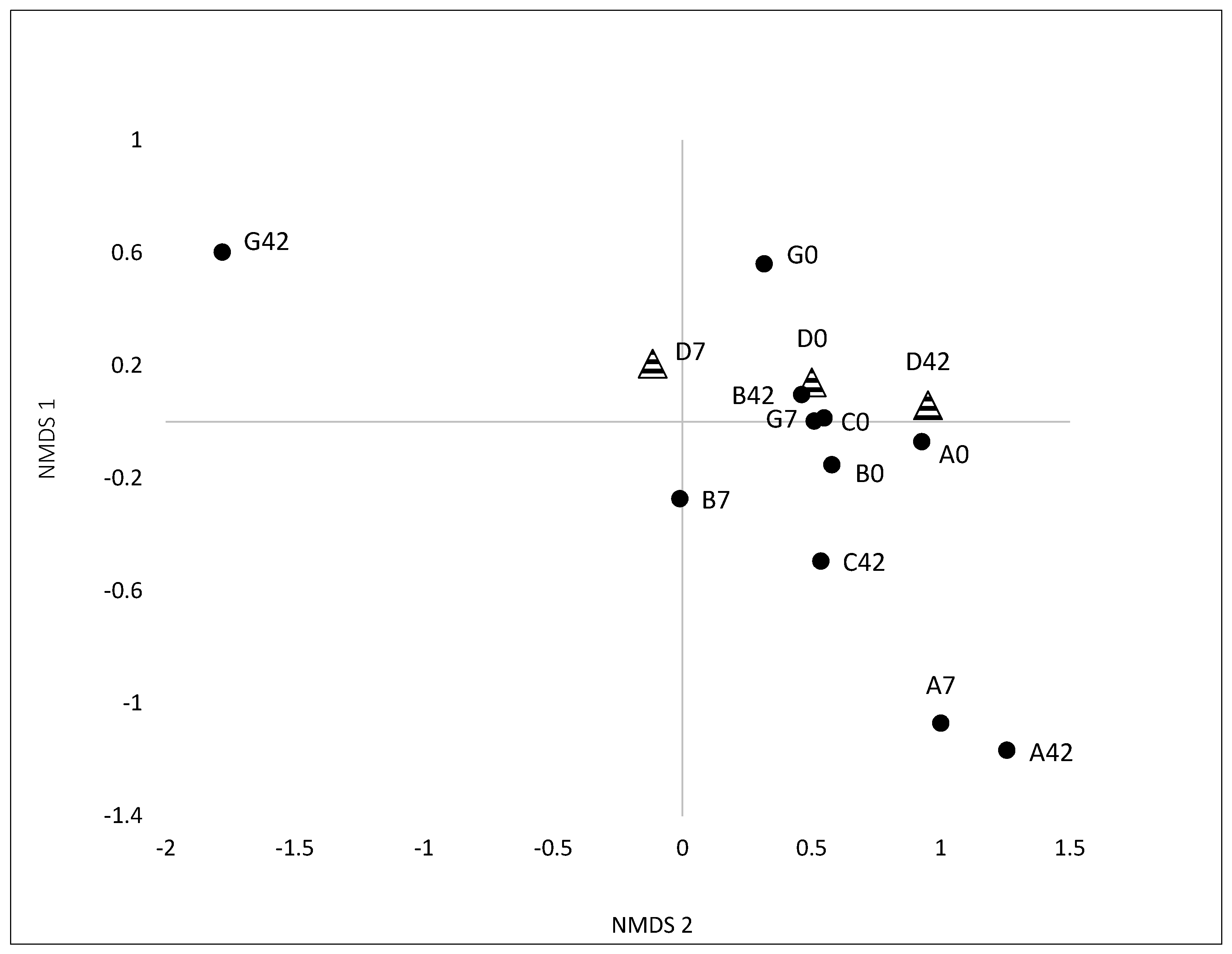
3.2.1. Variation and Diversity based on Analysis of T-RFLPs
3.2.2. Variation and Diversity based on NGS
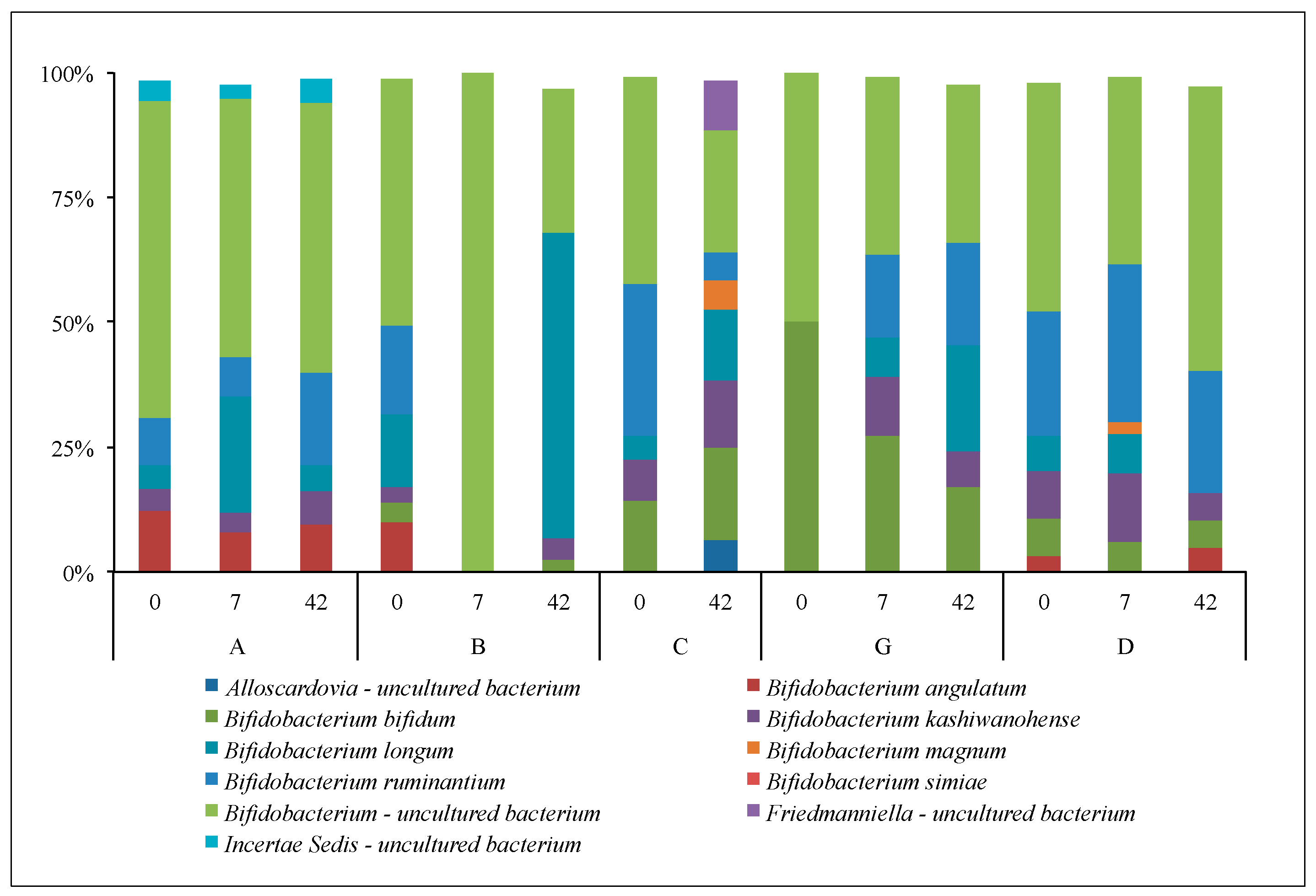
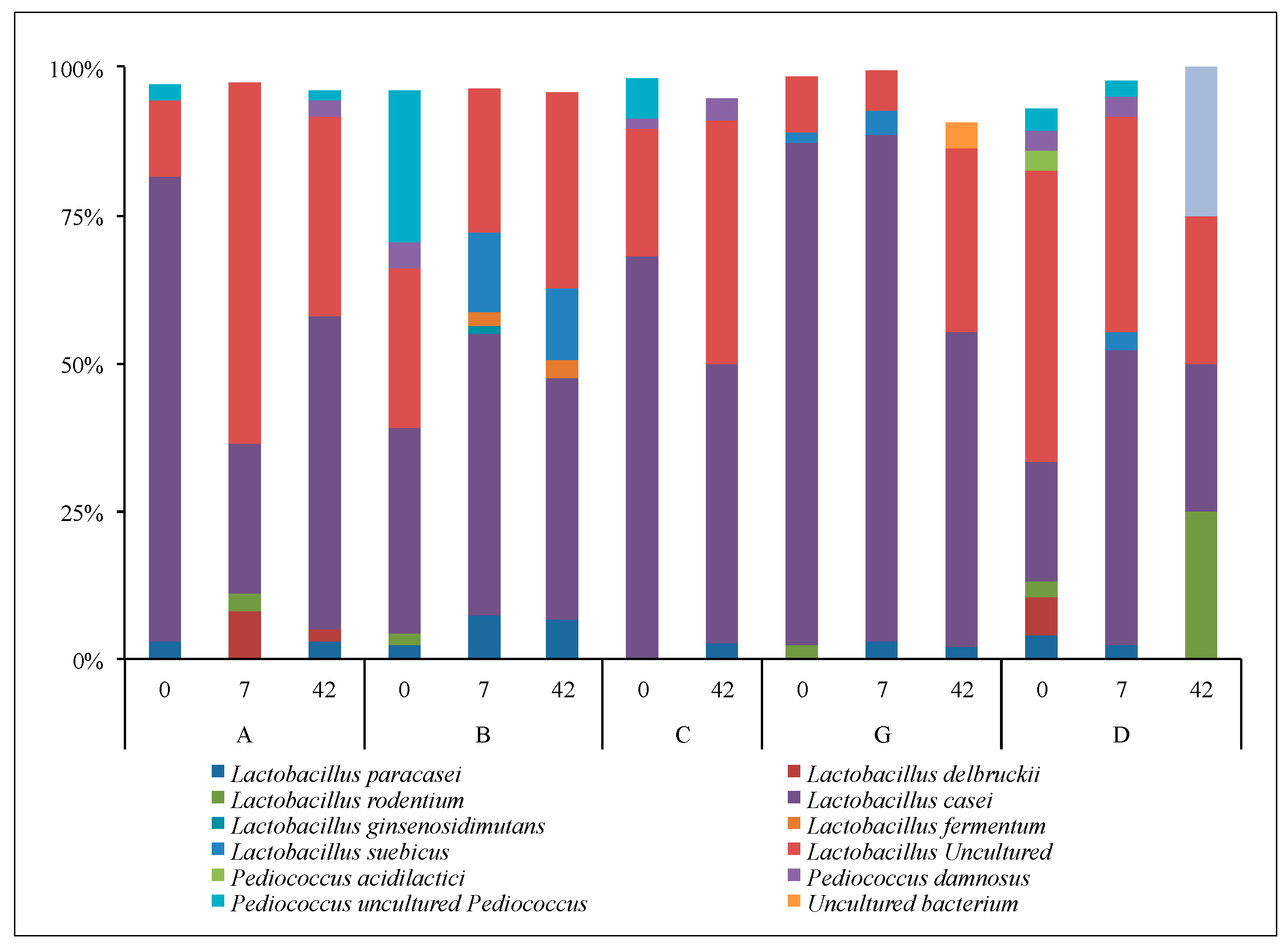
3.2.3. Lactobacilli and Bifidobacteria: Sanger Analyses
3.2.4. Lactobacilli and Bifidobacteria: NGS
3.3. Quantitative PCR of Lactobacillus 16s rDNA Genes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hamady, M.; Knight, R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009, 19, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Almada, C.; Amanda, C.; Ruiz, R.; de Souza, A. Characterization of the intestinal microbiota and its interaction with probiotics and health impacts. Appl. Microbiol. Biotechnol. 2015, 99, 4175–4199. [Google Scholar] [CrossRef] [PubMed]
- Rastall, R. Bacteria in the gut: Friends and foes and how to alter the balance. J. Nutr. 2004, 134, 2022–2026. [Google Scholar]
- Filteau, M.; Matamoros, S.; Savard, P.; Roy, D. Molecular monitoring of fecal microbiota in healthy adults following probiotic yogurt intake. PharmaNutrition 2013, 1, 123–129. [Google Scholar] [CrossRef]
- Kato-Kataoka, A.; Nishida, K.; Takada, M.; Kawai, M.; Kikuchi-Hayakawa, H. Fermented milk containing Lactobacillus casei strain Shirota preserves the diversity of the gut microbiota and relieves abdominal dysfunction in healthy medical students exposed to academic stress. Appl. Environ. Microbiol. 2016, 82, 3649–3658. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.; Antunes, L.; Finlay, B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E. Gut bacteria in health and disease. J. Gastroenterol. Hepatol. 2013, 9, 560–569. [Google Scholar]
- Preidis, G.; Versalovic, J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: Gastroenterology enters the metagenomics era. Gastroenterology 2009, 136, 2015–2031. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.; Lewis, J.; Hodgson, J.; Zhu, K.; Dhaliwal, S.; Thompson, P.; Prince, R. Association between yogurt, milk, and cheese consumption and common carotid artery intima-media thickness and cardiovascular disease risk factors in elderly women. Am. J. Clin. Nutr. 2011, 94, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Margolis, K.; Wei, F.; de Boer, I.; Howard, B.; Liu, S.; Manson, J. A diet high in low-fat dairy products lowers diabetes risk in postmenopausal women. J. Nutr. 2011, 141, 1969–1974. [Google Scholar] [CrossRef] [PubMed]
- Daliri, E.; Lee, N. New perspectives on probiotics in health and disease. Food Sci. Hum. Wellness 2015, 4, 56–65. [Google Scholar] [CrossRef]
- Yacoub, R.; Deepak, K.; Shanti, P.; Simoes, D.; Girish, N.; John, C. Association between probiotic and yogurt consumption and kidney disease: Insights from NAHES. Nutr. J. 2016, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Uyeno, Y. Impact of consumption of probiotic Lactobacilli-containing yogurt on microbial composition in human feces. Int. J. Food Microbiol. 2008, 122, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bor-Shyang, S. Probiotics-containing yogurts suppress Helicobacter pylori load and modify immune response and intestinal microbiota in the Helicobacter pylori-infected children. Helicobacter 2012, 17, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Maukonen, J.; Matto, J.; Suihko, M.; Saarela, M. Intra-individual diversity and similarity of salivary and faecal microbiota. J. Med. Microbiol. 2008, 57, 1560–1568. [Google Scholar] [CrossRef] [PubMed]
- Schütte, U.; Zaid Abdo, M.; Bent, S.; Shyu, C.; Williams, C.; Pierson, J.; Forney, L. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 2008, 80, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jang-Seu, K.; Pei-Yuan, Q. Microbial diversity in polluted harbor sediments I: Bacterial community assessment based on four clone libraries of 16S rDNA. Estuar. Coast. Shelf Sci. 2008, 76, 668–681. [Google Scholar] [CrossRef]
- Walter, J.; Hertel, C.; Tannock, G.; Lis, C.; Munro, K.; Hammes, W. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2001, 67, 2578–2585. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Miyamoto, Y.; Takada, T.; Matsumoto, K. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 2002, 68, 5445–5451. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Nei, M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004, 5, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Holland, M. Non Metric Multidimensional Scaling (MDS) University of Georgia, Department of Geology 2008. Available online: http://strata.uga.edu/software/pdf/mdsTutorial.pdf (accessed in November 2014).
- Caporaso, J.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F. The SILVA Ribosomal RNA Gene Database Project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, A. Lactobacillus acidophilus, Bifidobacterium lactis and Lactobacillus F19 prevent antibiotic-associated ecological disturbances of Bacteroides fragilis in the intestine. J. Antimicrob. Chemother. 2003, 52, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; van Hylckama Vlieg, J. Fate, activity and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015, 23, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Cormier, H.; Thifaul, E.; Garneau, V.; Tremblay, A.; Drapeau, V.; Perusse, L.; Vohl, M. Association between yogurt consumption, dietary patterns, and cardio-metabolic risk factors. Eur. J. Nutr. 2016, 55, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Margolis, K.; Wei, F.; de Boer, I.; Howard, B.; Liu, S.; Manson, J.E.; Mossavar-Rahmani, Y.; Phillips, L.S.; Shikany, J.M.; Tinker, L.F.; et al. A diet high in low-fat dairy products lowers diabetes risk in postmenopausal women. J. Nutr. 2011, 141, 1969–1974. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Livingston, K.; Fox, C.; Meigs, J.; Jacques, P. Yogurt consumption is associated with better diet quality and metabolic profile in American men and women. Nutr. Res. 2013, 33, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Ortinau, C.; Culp, J.; Hoertel, H.; Douglas, S.; Leidy, H. The effects of increased dietary protein yogurt snack in the afternoon on appetite control and eating initiation in healthy women. Nutr. J. 2013, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.; Lewis, J.; Hodgson, J.; Zhu, K.; Dhaliwal, S.; Thompson, P.; Prince, R. Association between yogurt, milk, and cheese consumption and common carotid artery intima-media thickness and cardiovascular disease risk factors in elderly women. Am. J. Clin. Nutr. 2011, 94, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Sayon-Orea, C.; Bes-Rastrollo, M.; Marti, A.; Pimenta, A.; Martin-Calvo, N.; Martinez-Gonzalez, M. Association between yogurt consumption and the risk of metabolic syndrome over 6 years in the SUN study. BMC Public Health 2015, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Heilig, H.; Zoetendal, E.; Vaughan, E.; Marteau, P.; Akkermans, P.; de Vos, W. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 2002, 68, 114–123. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Maurice, C.; Carmody, R.; Gootenberg, D. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omar, J.; Chan, Y.; Jones, M.; Prakash, S.; Jones, P. Lactobacillus fermentum and Lactobacillus amylovorus as probiotics alter body adiposity and gut microflora in healthy persons. J. Funct. Foods 2013, 5, 116–123. [Google Scholar] [CrossRef]
- Fouhy, F.; Clooney, A.; Stanton, C.; Claesson, M.; Cotter, P. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016, 16, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spellerberg, I.; Fedor, P. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the “Shannon-Wiener” index. Glob. Ecol. Biogeogr. 2003, 12, 177–179. [Google Scholar] [CrossRef]
- Karlsson, C.; Ahrne, S.; Molin, G.; Berggren, A.; Palmquist, I.; Fredrikson, G.; Jeppsson, B. Probiotic therapy to men with incipient arteriosclerosis initiates increased bacterial diversity in colon: A randomized controlled trial. Atherosclerosis 2010, 208, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Sankar, S.; Lagier, J.; Pontarotti, P.; Raoult, D.; Fournier, P. The human gut microbiome, a taxonomic conundrum. Syst. Appl. Microbiol. 2015, 38, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Suda, W.; Kim, S.; Oshima, K.; Fukuda, S.; Ohno, H.; Morita, H.; Hattori, M. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 2013, 20, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Kelso, J. High-throughput DNA sequencing—Concepts and limitations. Bioessays 2010, 32, 524–536. [Google Scholar] [CrossRef] [PubMed]
| Target | Primers | Sequence (5'-3') | PCR Settings | Ref. | Used for |
|---|---|---|---|---|---|
| Eubacteria | 8F 1492R (~1400 bp) | AGAGTTTGATCATGGCTCAG GGCTACCTTGCCACGACTTC | 94 °C 3 min 94 °C 30 s 53 °C 1 min (35×) 72 °C 90 s 72 °C 7 min | [18] | T-RFLP, NGS |
| Lactobacillus | Lac 1 Lac 2 (380 bp) | AGCAGTAGGGAATCTTCCA ATTYCACCGCTACACATG | 94°C 3 min 94°C 30 s 53°C 1 min (35×) 68°C 1 min 68°C 7 min | [19] | NGS, Sanger, qPCR |
| Bifidobacteria | G-Bifid-F G-Bifid-R (549–563 bp) | CTCCTGGAAACGGGTGG CGTGTTCTTCCCGATATCTACA | 95°C 5 min 95°C 30 s 55°C 20 s (30×) 72°C 30 s 72°C 5 min | [20] | NGS, Sanger, qPCR |
| Days | Subjects | ||||||
|---|---|---|---|---|---|---|---|
| A | B | C | F | G | D | ||
| 0 | 11 | 12 | 18 | 8 | 12 | 15 | |
| 7 | 13 | 13 | 16 | 17 | 10 | 15 | |
| 14 | 12 | 18 | 17 | 15 | 11 | 15 | |
| 21 | 10 | 16 | 15 | 12 | 13 | 14 | |
| 28 | 14 | 17 | 20 | 14 | 13 | 12 | |
| 35 | 14 | 15 | 15 | 15 | 14 | 10 | |
| 42 | 15 | 15 | 17 | 15 | 10 | 11 | |
| Day | A | B | C | F | G | D |
|---|---|---|---|---|---|---|
| 0 | 2.49 | 2.63 | 3.06 | 2.25 | 2.32 | 2.77 |
| 7 | 2.72 | 2.86 | 3.18 | 2.44 | 2.36 | 2.98 |
| 14 | 2.78 | 3.11 | 3.33 | 3.18 | 2.39 | 3.07 |
| 21 | 2.64 | 3.00 | 3.02 | 2.35 | 2.86 | 2.80 |
| 28 | 2.84 | 3.38 | 3.32 | 2.87 | 2.71 | 2.83 |
| 35 | 2.56 | 2.91 | 3.00 | 2.83 | 2.60 | 2.48 |
| 42 | 2.38 | 3.11 | 3.13 | 2.73 | 2.67 | 2.99 |
| Eubacteria | ||||
|---|---|---|---|---|
| Subject | H’ | Chao1 | ||
| Day 0 | Day 42 | Day 0 | Day 42 | |
| A | 5.35 | NA | 89.37 | NA |
| B | 5.33 | 5.36 | 95.77 | 103.19 |
| C | 5.33 | 5.25 | 94.87 | 95.43 |
| G | 5.33 | 3.48 | 99.53 | 48.63 |
| D | 5.31 | 5.41 | 96.50 | 119.04 |
| Subject | Day 0 | Day 42 | ||
|---|---|---|---|---|
| Bifidobacterium | Lactobacillus | Bifidobacterium | Lactobacillus | |
| B | B. longum 19% B. pseudocatenulatum 81% | L. iners 27% L. casei 73% | B. longum 100% | L. casei 95% L. rhamnosus 5% |
| C | B. longum 21% B. bifidum 44% B. adolescentis 24% B. pseudocatenulatum 10% | L. gasseri 45% L. crispatus 20% L. casei 10% Pediococcus stilesii 20% | B. longum 17% B. bifidum 17% B. adolescentis 62% Alloscardovia omnicolens 4% | L. casei 26% L. rhamnosus 26% L. reuteri 26% L. ruminis 22% |
| D | B. longum 47% B. adolescentis 44% B. pseudocatenulatum 9% | L. gasseri 8% L. crispatus 4% L. sakei 4% L. casei 13% L. rhamnosus 29% L. plantarum 8% Dolosigranulum pigrum 4% Granulicatella adiacens 4% Pediococcus stilesii 25% | B. bifidum 8% B. adolescentis 41% B. pseudocatenulatum 51% | L. gasseri 69% L. delbrueckii 9% L. homohiochii 6% Weissella paramesenteroides 17% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lisko, D.J.; Johnston, G.P.; Johnston, C.G. Effects of Dietary Yogurt on the Healthy Human Gastrointestinal (GI) Microbiome. Microorganisms 2017, 5, 6. https://doi.org/10.3390/microorganisms5010006
Lisko DJ, Johnston GP, Johnston CG. Effects of Dietary Yogurt on the Healthy Human Gastrointestinal (GI) Microbiome. Microorganisms. 2017; 5(1):6. https://doi.org/10.3390/microorganisms5010006
Chicago/Turabian StyleLisko, Daniel J., G. Patricia Johnston, and Carl G. Johnston. 2017. "Effects of Dietary Yogurt on the Healthy Human Gastrointestinal (GI) Microbiome" Microorganisms 5, no. 1: 6. https://doi.org/10.3390/microorganisms5010006





