Tellurite and Tellurate Reduction by the Aerobic Anoxygenic Phototroph Erythromonas ursincola, Strain KR99 Is Carried out by a Novel Membrane Associated Enzyme
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tellurite Reductase Purification and Characterization
2.2. Enzyme Properties and Kinetics
2.3. Mass Spectrometry
3. Results
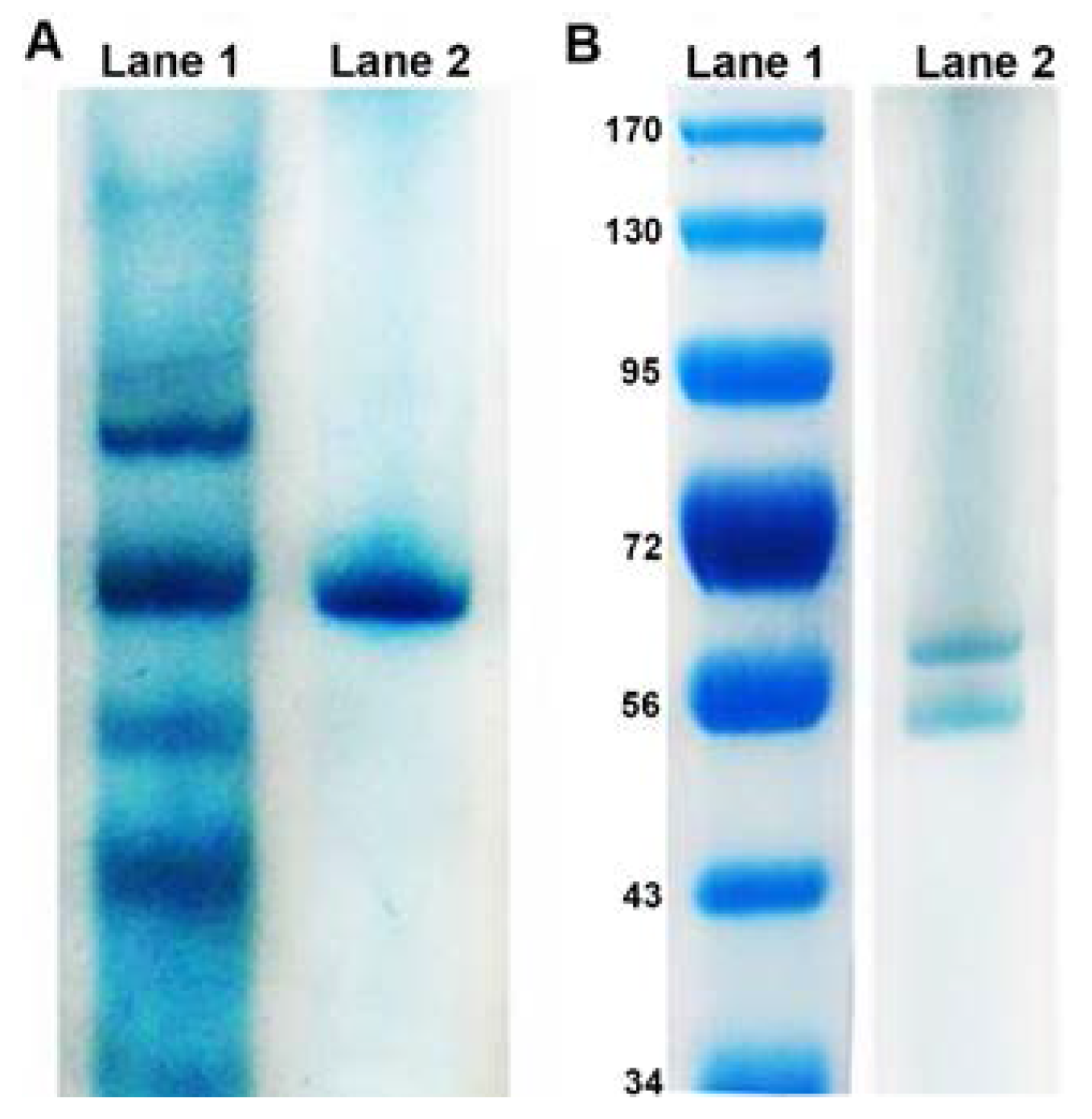
3.1. Physical Characteristics of Reductase
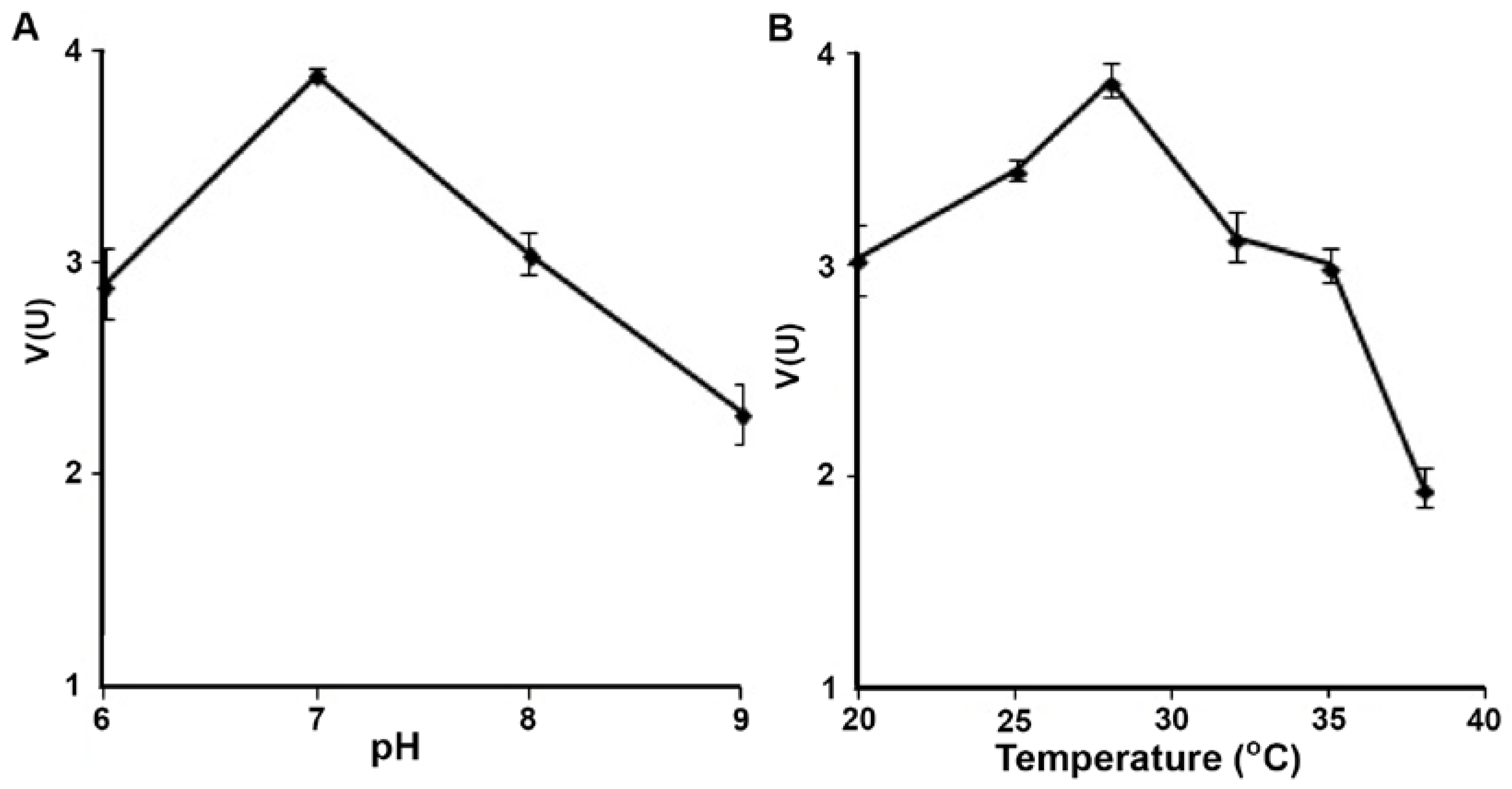
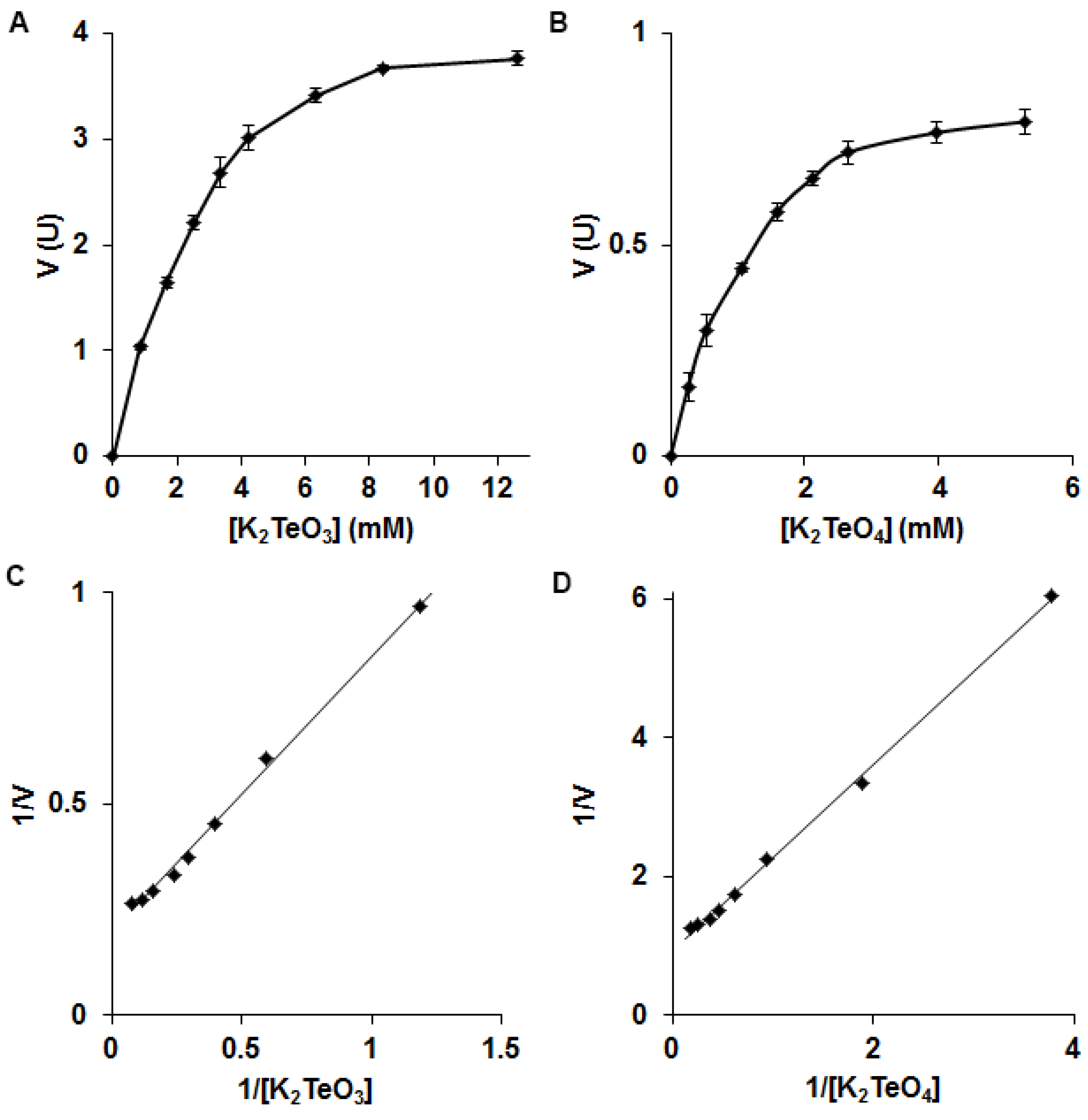
3.2. Biochemistry of Reductase
3.3. Mass Spectrometry Analysis
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yurkov, V.; Jappe, J.; Vermeglio, A. Tellurite resistance and reduction by obligately aerobic photosynthetic bacteria. Appl. Environ. Microbiol. 1996, 62, 4195–4198. [Google Scholar] [PubMed]
- Taylor, D. Bacterial tellurite resistance. Trends Microbiol. 1999, 7, 111–115. [Google Scholar] [CrossRef]
- Lloyd, J.; Mabbett, A.; Williams, D.; Macaskie, L. Metal reduction by sulfate-reducing bacteria: Physiological diversity and metal specificity. Hydrometallurgy 2001, 59, 327–337. [Google Scholar] [CrossRef]
- Avazeri, C.; Turner, J.; Weiner, J.; Giordano, G.; Vermeglio, A. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology 1997, 143, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Sabaty, M.; Avazeri, C.; Pignol, D.; Vermeglio, A. Characterization of the reduction of selenate and tellurite by nitrate reductases. Appl. Environ. Microbiol. 2001, 67, 5122–5126. [Google Scholar] [CrossRef] [PubMed]
- Borsetti, F.; Francia, F.; Turner, R.; Zannoni, D. The thiol:disulfide oxidoreductase DsbB mediates the oxidizing effects of the toxic metalloid tellurite (TeO32−) on the plasma membrane redox system of the facultative phototroph Rhodobacter capsulatus. J. Bacteriol. 2007, 189, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Guzzo, J.; Dubow, M. A novel selenite- and tellurite-inducible gene in Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 4972–4978. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.; Lou, Y.; Chen, C. NMR solution of KP-TerB, a tellurite-resistance protein from Klebsiella pneumoniae. Protein Sci. 2008, 17, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Chiong, M.; Gonzalez, E.; Barra, R.; Vasquez, C. Purification and biochemical characterization of tellurite-reducing activities from Thermus thermophilus HB8. J. Bacteriol. 1988, 170, 3269–3273. [Google Scholar] [CrossRef] [PubMed]
- Kabiri, M.; Amoozegar, M.; Tabebordbar, M.; Gilany, K.; Salekdeh, G. Effects of selenite and tellurite on growth, physiology, and proteome of a moderately halophilic bacterium. J. Proteome Res. 2009, 8, 3098–3108. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, H.; Saavedra, C.; Loyola, C.; Pichuantes, S.; Vasquez, C. Biochemical characterization of tellurite-reducing activities of Bacillus stearothermophilus V. Res. Microbiol. 1998, 149, 389–397. [Google Scholar] [CrossRef]
- Etezad, S.; Khajeh, K.; Soudi, M.; Ghazvini, P.; Dabirmanesh, B. Evidence on the presence of two distinct enzymes responsible for the reduction of selenate and tellurite in Bacillus sp. STG-83. Enzyme Microb. Technol. 2009, 45, 1–6. [Google Scholar] [CrossRef]
- Moore, M.; Kaplan, S. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: Characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J. Bacteriol. 1992, 17, 1505–1514. [Google Scholar] [CrossRef]
- Moore, M.; Kaplan, S. Members of the family Rhodospirillaceae reduce heavy-metal oxyanions to maintain redox poise during photosynthetic growth. ASM News 1994, 60, 17–23. [Google Scholar]
- Borghese, R.; Zannoni, D. Acetate permease (ActP) is responsible for tellurite (TeO32−) uptake and resistance in cells of the facultative phototroph Rhodobacter capsulatus. Appl. Environ. Microbiol. 2010, 76, 942–944. [Google Scholar] [CrossRef] [PubMed]
- Borghese, R.; Marchetti, D.; Zannoni, D. The highly toxic oxyanion tellurite (TeO32−) enters the phototrophic bacterium Rhodobacter capsulatus via an as yet uncharacterized monocarboxylate transport system. Arch. Microbiol. 2008, 189, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Borghese, R.; Canducci, L.; Musiani, F.; Cappelletti, M.; Ciurli, S.; Turner, R.; Zannoni, D. On the role of a specific insert in acetate permeases (ActP) for tellurite uptake in bacteria: Functional and structural studies. J. Inorg. Biochem. 2016. [Google Scholar] [CrossRef] [PubMed]
- Tomás, J.; Kay, W. Tellurite susceptibility and non-plasmid-mediated resistance in Escherichia coli. Antimicrob. Agents Chemother. 1986, 30, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Ollivier, P.; Bahrou, A.; Marcus, S.; Cox, T.; Church, T.; Hanson, T. Volatilization and precipitation of tellurium by aerobic tellurite-resistant marine microbes. Appl. Environ. Microbiol. 2008, 74, 7163–7173. [Google Scholar] [CrossRef] [PubMed]
- Csotonyi, J.; Stackebrandt, E.; Yurkov, V. Anaerobic respiration on tellurate and other metalloids in bacteria from hydrothermal vent fields in the eastern Pacific Ocean. Appl. Environ. Microbiol. 2006, 72, 4950–4956. [Google Scholar] [CrossRef] [PubMed]
- DeMoll-Decker, H.; Macy, J. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch. Microbiol. 1993, 160, 241–247. [Google Scholar]
- Laverman, A.; Blum, J.; Schaefer, J.; Phillips, E.; Lovley, D.; Oremland, R. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Appl. Environ. Microbiol. 1995, 61, 3556–3561. [Google Scholar] [PubMed]
- Wood, J. Biological cycles for toxic elements in the environment. Science 1974, 4129, 1049–1052. [Google Scholar] [CrossRef]
- Macy, J.; Lawson, S.; DeMoll-Decker, H. Bioremediation of selenium oxyanions in San Joaquin drainage water using Thauera selenatis in a biological reactor system. Appl. Environ. Microbiol. 1993, 40, 588–594. [Google Scholar] [CrossRef]
- Cantafio, A.; Hagen, K.; Lewis, G.; Bledsoe, T.; Nunan, K.; Macy, J. Pilot-scale selenium bioremediation of San Joaquin drainage water with Thaurea selenatis. Appl. Environ. Microbiol. 1996, 62, 3298–3303. [Google Scholar] [PubMed]
- Rajwade, J.; Paknikar, K. Bioreduction of tellurite to elemental tellurium by Pseudomonas mendocina MCM B-180 and its practical application. Hydrometallurgy 2003, 71, 243–248. [Google Scholar] [CrossRef]
- Yurkov, V.; Csotonyi, J. Aerobic anoxygenic phototrophs and heavy metalloid reducers from extreme environments. Recent Res. Dev. Bacteriol. 2003, 1, 247–300. [Google Scholar]
- Maltman, C.; Yurkov, V. The effect of tellurite on highly resistant freshwater aerobic anoxygenic phototrophs and their strategies for reduction. Microorganisms 2015, 3, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Yurkov, V.; Gorlenko, V. Erythrobacter sibiricus sp. nov., a new freshwater aerobic bacterial species containing bacteriochlorophyll A. Mikrobiologiya 1990, 59, 85–89. [Google Scholar]
- Yurkov, V.; Gorlenko, V. New species of aerobic bacteria from the genus Erythromicrobium containing bacteriochlorophyll A. Mikrobiologiya 1992, 61, 163–168. [Google Scholar]
- Yurkov, V.; Gorlenko, V.; Kompantseva, E. A new genus of orange-coloured bacteria containing bacteriochlorophyll A; Erythromicrobium gen. nov. Mikrobiologiya 1992, 61, 256–260. [Google Scholar]
- Yurkov, V.; Lysenko, A.; Gorlenko, V. Hybridization analysis of the classification of bacteriochlorophyll a-containing freshwater aerobic bacteria. Mikrobiologiya 1991, 60, 362–366. [Google Scholar]
- Yurkov, V.; Stackebrandt, E.; Holmes, A.; Fuerst, J.; Hugenholtz, P.; Golecki, J.; Gad’on, N.; Gorlenko, V.; Kompantseva, E.; Drews, G. Phylogenetic positions of novel aerobic bacteriochlorophyll a-containing bacteria and description of Roseococcus thiosulfatophilus gen. nov., sp. nov., Erythromicrobium ramosum gen. nov., sp. nov., and Erythrobacter litoralis sp. nov. Int. J. Syst. Bacteriol. 1994, 44, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C. Staining of proteins on gels: Comparison of dyes and procedures. Methods Enzymol. 1983, 91, 236–247. [Google Scholar] [PubMed]
- Molina, R.; Burra, R.; Perez-Donoso, J.; Elias, A.; Munoz, C.; Montes, R.; Chasteen, T.; Vasquez, C. Simple, fast, and sensitive method for quantification of tellurite in culture media. Appl. Environ. Microbiol. 2010, 76, 4901–4904. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havliš, J.; Olse, J.; Mann, M. In gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Loboda, A.; Shevchenko, M.; Ens, W.; Standing, K. MALDI quadrupole time-of-flight mass spectrometry: A powerful tool for proteomic research. Anal. Chem. 2000, 72, 2132–2141. [Google Scholar] [CrossRef] [PubMed]
- Matrix Science Limited. Available online: http://www.matrixscience.com (accessed on 22 February 2016).
- Lima, A.; Siquerira, A.; Garcia, B.; Santos, S.; da Silva, F.; Lima, C.; Cardoso, J.; Vianez-Junior, J.; Nunes, M.; Goncalves, E. Draft genome sequence of Blastomonas sp. strain CACIA 14H2, a heterotrophic bacterium associated with cyanobacteria. Genome Announc. 2014, 2, e01200-13. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Hipp, M.; Bracher, A.; Hayer-Hartl, M.; Hartl, F. Molecular chaperone functions in protein folding and proteostasis. Ann. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Huberts, D.; van der Klei, I. Moonlighting proteins: An intriguing mode of multitasking. Biochem. Biophys. Acta 2010, 1803, 520–525. [Google Scholar] [CrossRef] [PubMed]
| Fraction | Activity (µM K2TeO3/min) | Total Protein (mg/L) | Specific Activity (µM K2TeO3/min/mg Protein) | Yield (%) | Fold Purification |
|---|---|---|---|---|---|
| Cell Lysate | 8.14 | 261 | 0.031 | 100 | 1 |
| Membranes | 2.25 | 31 | 0.073 | 27.64 | 2.36 |
| S200 fraction | 1.87 | 0.64 | 2.94 | 22.97 | 94.84 |
| Ion Exchange fraction | 1.31 | 0.28 | 4.71 | 16.09 | 151.94 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maltman, C.; Donald, L.J.; Yurkov, V. Tellurite and Tellurate Reduction by the Aerobic Anoxygenic Phototroph Erythromonas ursincola, Strain KR99 Is Carried out by a Novel Membrane Associated Enzyme. Microorganisms 2017, 5, 20. https://doi.org/10.3390/microorganisms5020020
Maltman C, Donald LJ, Yurkov V. Tellurite and Tellurate Reduction by the Aerobic Anoxygenic Phototroph Erythromonas ursincola, Strain KR99 Is Carried out by a Novel Membrane Associated Enzyme. Microorganisms. 2017; 5(2):20. https://doi.org/10.3390/microorganisms5020020
Chicago/Turabian StyleMaltman, Chris, Lynda J. Donald, and Vladimir Yurkov. 2017. "Tellurite and Tellurate Reduction by the Aerobic Anoxygenic Phototroph Erythromonas ursincola, Strain KR99 Is Carried out by a Novel Membrane Associated Enzyme" Microorganisms 5, no. 2: 20. https://doi.org/10.3390/microorganisms5020020





