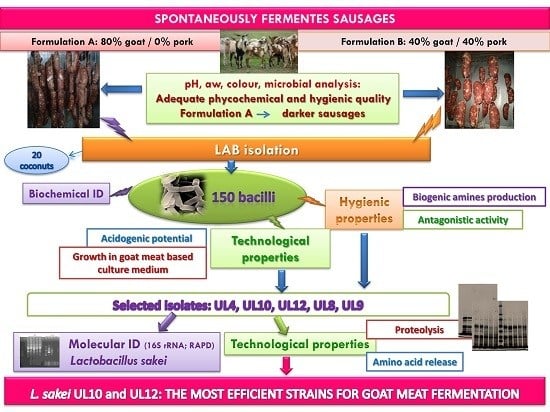
Adding Value to Goat Meat: Biochemical and Technological Characterization of Autochthonous Lactic Acid Bacteria to Achieve High-Quality Fermented Sausages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Fermented Sausage and Sampling
2.2. Determination of Water Activity (aw) and pH
2.3. Color Analysis
2.4. Microbiological Analysis and LAB Isolation
2.5. Phenotypic and Technological Characterization of LAB
2.6. Determination of Hygienic and Technological Properties of Fermented Sausage Isolates
2.6.1. Decarboxylase Activity
2.6.2. Screening for Antagonistic Activity
2.6.3. Lipolytic Activity
2.6.4. Proteolytic Activity
2.7. Growth, Adaption and Acidifying Ability in Meat Environments
2.7.1. Meat Model System and Culture Conditions
2.7.2. Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.7.3. Free Amino acid and Small Peptide Analysis
2.8. Genotypic Characterization of Isolates
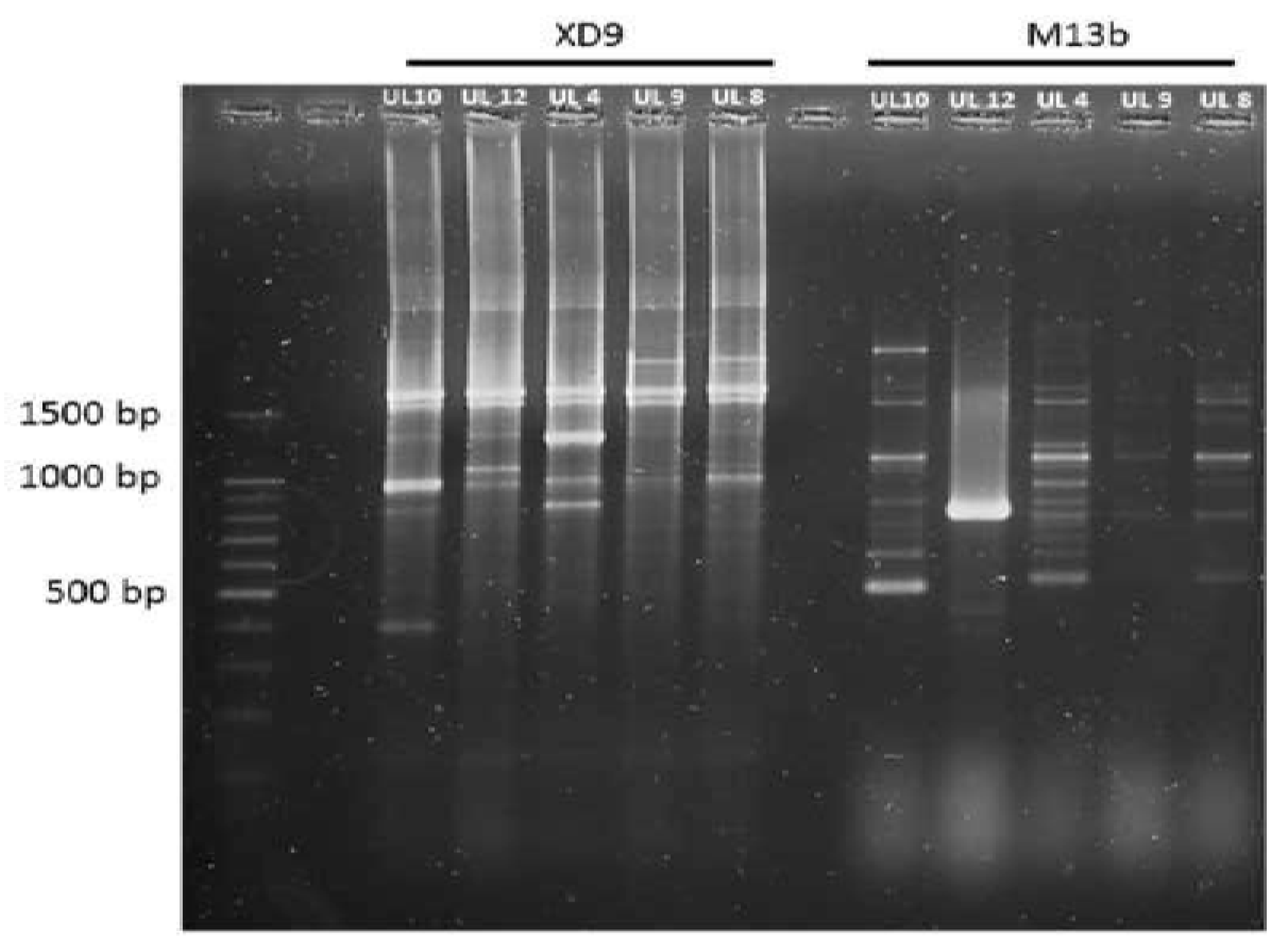
2.9. Random Amplified Polymorphic DNA-PCR (RAPD-PCR) Analysis
2.10. Data Analysis
3. Results and Discussion
3.1. Physicochemical Analysis of Fermented Sausages
3.1.1. pH and aw
3.1.2. Color
3.2. Microbial Analysis
3.2.1. Microorganisms with Technological Importance
3.2.2. Microorganisms Related to Food Spoilage and Food-Borne Illness
3.3. Phenotypic Characterization of Lactic Acid Bacteria Isolated from Goat-Meat Fermented Sausages
3.4. Hygienicand Technological Properties of LAB Isolated from Goat-Meat Fermented Sausages
3.4.1. Decarboxylase Activity
3.4.2. Adaption to Meat Environments
3.4.3. Ability to Hydrolyze Fats and Proteins from Meat
Lipolytic Activity
Proteolytic Activity
Free Amino Acid and Small Peptide Analysis
3.5. Genotypic Characterization of LAB Strains Isolated from Goat-Meat Fermented Sausages with the Highest Technological Potential
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Madruga, M.S.; Bressan, M.C. Goat meats: Description, rational use, certification, processing and technological developments. Small Rumin. Res. 2011, 98, 39–45. [Google Scholar] [CrossRef]
- Catan, A.; Degano, C.A.M. Composiciónbotánica de la dieta de caprinos en un bosque del Chaco semiárido (Argentina). Quebracho 2007, 14, 1851–3026. [Google Scholar]
- Madruga, M.S.; Torres, T.S.; Carvalho, F.F.; Queiroga, R.C.; Narain, N.; Garrutti, D.; Souza Neto, M.A.; Mattos, C.W.; Costa, R.G. Meat quality of Moxotó and Canindé goats as affected by two levels of feeding. Meat Sci. 2008, 80, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Nassu, R.T.; Gonçalves, L.A.G.; Beserra, F.J.; Feitosa, T. Estudo das características físico-químicas, microbiológicas e sensoriais de embutidos fermentados tipo salame formulados com diferentes proporçöes de carne caprina e suína. Bol. Centro Pesqui. Process. Aliment. 2001, 19, 243–256. [Google Scholar] [CrossRef]
- Talon, R.; Leroy, S.; Lebert, I.; Giammarinaro, P.; Chacornac, J.P.; Latorre-Moratalla, M.; Vidal-Carou, C.; Zanardi, E.; Conter, M.; Lebecque, A. Safety improvement and preservation of typical sensory qualities of traditional dry fermented sausages using autochthonous starter cultures. Int. J. Food Microbiol. 2008, 126, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Lücke, F.K. Utilization of microbes to process and preserve meat. Meat Sci. 2000, 56, 105–115. [Google Scholar] [CrossRef]
- Talon, R.; Leroy-Sétrin, S.; Fadda, S. Bacterial starters involved in the quality of fermented meat products. In Research Advances in the Quality of Meat and Meat Products, 1st ed.; Toldrá, F., Ed.; Research Singpost: Trivandrum, India, 2002; Volume 1, pp. 175–191. ISBN 81-7736-125-2. [Google Scholar]
- Fadda, S.; Lopez, C.; Vignolo, G. Role of lactic acid bacteria during meat conditioning and fermentation: Peptides generated as sensorial and hygienic biomarkers. Meat Sci. 2010, 86, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Ravyts, F.; Steen, L.; Goemaere, O.; Paelinck, H.; De Vuyst, L.; Leroy, F. The application of staphylococci with flavour-generating potential is affected by acidification in fermented dry sausages. Food Microbiol. 2010, 27, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Perez-Alvarez, J.A.; Sayas-Barbera, M.E.; Fernandez-Lopez, J.; Aranda-Catala, V. Physicochemical characteristics of Spanish-type dry-cured sausage. Food Res. Int. 1999, 32, 599–607. [Google Scholar] [CrossRef]
- Food and Drug Administration (FDA). Bacteriological Analytical Manual, 7th ed.; Association of Official Analytical, Chemists: Arlington, VA, USA, 1992; pp. 51–70. ISBN 0-935584-49-8.
- International Comission on Microbiological Specifications for Foods ((ICMSF). Microrganismos de los Alimentos 1: Técnicas de Análisis Microbiológico; Editorial Acribia: Zaragoga, España, 1982; Volume 2, p. 431. [Google Scholar]
- Xanthopoulos, V.; Polychroniadou, A.; Litopoulou-Tzanetaki, E.; Tzanetakis, N. Characteristics of Anevato Cheese made from Raw or Heat-treated Goat Milk Inoculated with a Lactic Starter. Food Sci. Technol. Int. 2000, 33, 483–488. [Google Scholar] [CrossRef]
- Drosinos, E.H.; Mataragas, M.; Vesković-Moračanin, S.; Gasparik-Reichardt, J.; Hadžiosmanović, M.; Alagić, D. Quantifying nonthermal inactivation of Listeria monocytogenes in European fermented sausages using bacteriocinogenic lactic acid bacteria or their bacteriocins: A case study for risk assessment. J. Food Prot. 2006, 69, 2648–2663. [Google Scholar] [CrossRef] [PubMed]
- Joosten, H.M.L.J.; Northolt, M.D. Detection, growth, and amine-producing capacity of lactobacilli in cheese. Appl. Environ. Microbiol. 1989, 55, 2356–2359. [Google Scholar] [PubMed]
- Saavedra, L.; Castellano, P.; Sesma, F. Purification of Bacteriocins Produced by Lactic Acid Bacteria. In Public Health Microbiology: Methods and Protocols; Spencer, J.F.T., de Spencer, A.L.R., Eds.; Humana Press: Totowa, NJ, USA, 2004; Volume 30, pp. 331–336. ISBN 1-58829-117-0. [Google Scholar]
- Medina, R.B.; Katz, M.B.; González, S. Differentiation of lactic acid bacteria strains by postelectrophoretic detection of esterases. In Public Health Microbiology: Methods and Protocols; Spencer, J.F.T., de Spencer, A.L.R., Eds.; Humana Press: Totowa, NJ, USA, 2004; Volume 268, pp. 459–463. ISBN 1-58829-117-0. [Google Scholar]
- Fadda, S.; Sanz, Y.; Vignolo, G.; Aristoy, M.C.; Oliver, G.; Toldra, F. Characterization of muscle sarcoplasmic and myofibrillar protein hydrolysis caused by Lactobacillus plantarum. Appl. Environ. Microbiol. 1999, 65, 3540–3546. [Google Scholar] [PubMed]
- Fadda, S.; Vignolo, G.; Holgado, A.P.; Oliver, G. Proteolytic activity of Lactobacillus strains isolated from dry fermented sausages on muscle sarcoplasmic proteins. Meat Sci. 1998, 49, 11–18. [Google Scholar] [CrossRef]
- Nassu, R.T.; Gonçalves, L.A.G.; Beserra, F.J. Efeito do teor de gordura nas características químicas e sensoriais de embutido fermentado de carne de caprinos. Pesqui. Agropecu. Bras. 2002, 37, 1169–1173. [Google Scholar] [CrossRef]
- Church, F.C.; Swaisgood, H.E.; Porter, D.H.; Catignani, G.L. Spectrophotometric assay using O-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 1983, 66, 1219–1227. [Google Scholar] [CrossRef]
- Pospiech, A.; Neumann, B. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 1995, 11, 217–218. [Google Scholar] [CrossRef]
- Fontana, C.; Cocconcelli, P.; Vignolo, G. Monitoring the bacterial population dynamics during fermentation of artisanal Argentinean sausages. J. Food Microbiol. 2005, 103, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Olivares, A.; Navarro, J.L.; Salvador, A.; Flores, M. Sensory acceptability of slow fermented sausages based on fat content and ripening time. Meat Sci. 2010, 86, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Casquete, R.; Benito, M.J.; Martín, A.; Ruiz-Moyano, S.; Hernández, A.; Córdoba, M.G. Effect of autochthonous starter cultures in the production of “salchichón”, a traditional Iberian dry-fermented sausage, with different ripening processes. Food Sci. Technol. 2011, 44, 1562–1571. [Google Scholar] [CrossRef]
- Vignolo, G.; Fontana, C.; Fadda, S. Semidry and dry fermented sausages. In Handbook of Meat Processing; Toldrá, F., Ed.; Willey-Blackwell: Iowa, IA, USA, 2010; pp. 379–398. ISBN 978-0-8138-2182-5. [Google Scholar]
- Marty, E.; Buchs, J.; Eugster-Meier, E.; Lacroix, C.; Meile, L. Identification of staphylococci and dominant lactic acid bacteria in spontaneously fermented Swiss meat products using PCR–RFLP. Food Microbiol. 2012, 29, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Janssens, M.; Myter, N.; De Vuyst, L.; Leroy, F. Species diversity and metabolic impact of the microbiota are low in spontaneously acidified Belgian sausages with an added starter culture of Staphylococcus carnosus. Food Microbiol. 2012, 29, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Møller, J.K.S.; Jongberg, S.; Skibsted, L.H. Color. In Handbook of Fermented Meat and Poultry, 2nd ed.; Toldrá, F., Hui, Y.H., Astiasarán, I., Sebranek, J.G., Talon, R., Eds.; Wiley Blackwell: Chichester, UK, 2007; pp. 203–216. ISBN 978-0-8138-1477-3/2007. [Google Scholar]
- Stajić, S.; Perunović, M.; Stanišić, N.; Žujovi, M.; Živković, D. Sucuk (Turkish-style dry-fermented sausage) quality as an influence of recipe formulation and inoculation of starter cultures. J. Food Process. Preserv. 2013, 37, 870–880. [Google Scholar] [CrossRef]
- Teixeira, A.; Pereira, E.; Rodrigues, E.S. Goat meat quality. Effects of salting, air-drying and ageing processes. Small Rumin. Res. 2011, 98, 55–58. [Google Scholar] [CrossRef]
- Mancini, R.A.; Hunt, M. Current research in meat color. Meat Sci. 2005, 71, 100–121. [Google Scholar] [CrossRef] [PubMed]
- Paleari, M.A.; Bersani, C.; Vittorio, M.M.; Beretta, G. Effect of curing and fermentation on the microflora of meat of various animal species. Food Control 2002, 13, 195–197. [Google Scholar] [CrossRef]
- Casaburi, A.; Di Monaco, R.; Cavella, S.; Toldrá, F.; Ercolini, D.; Villani, F. Proteolytic and lipolytic starter cultures and their effect on traditional fermented sausages ripening and sensory traits. Food Microbiol. 2008, 25, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Fernández-López, J.; Sendra, E.; Sayas-Barberá, E.; Navarro, C.; Pérez-Alvarez, J.A. Physico-chemical and microbiological profiles of “salchichón” (Spanish dry-fermented sausage) enriched with orange fiber. Meat Sci. 2008, 80, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Urso, R.; Rantsiou, K.; Cantoni, C.; Comi, G. Dynamics and characterization of yeasts during natural fermentation of Italian sausages. FEMS Yeast Res. 2006, 6, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.; Dura, M.A.; Marco, A.; Toldrá, F. Effect of Debaryomyces spp. On aroma formation and sensory quality of dry-fermented sausages. Meat Sci. 2004, 68, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Martín, B.; Garriga, M.; Hugas, M.; Bover-Cid, S.; Veciana-Nogués, M.T.; Aymerich, T. Molecular, technological and safety characterization of Gram-positive catalase-positive cocci from slightly fermented sausages. Int. J. Food Microbiol. 2006, 107, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Doulgeraki, A.I.; Paramithiotis, S.; Kagkli, D.M.; Nychas, G.J.E. Lactic acid bacteria population dynamics during minced beef storage under aerobic or modified atmosphere packaging conditions. Food Microbiol. 2010, 27, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Cordero, M.; Zamalacárregui, J. Caracterization of Micrococcaceae isolates from salt used for Spanish dry-cured ham. Lett. Appl. Microbiol. 2000, 31, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Vignolo, G.; Fontana, C.; Cocconcelli, P.S. New approaches for the study of lactic acid bacteria biodiversity: A focus on meat ecosystems. In Biotechnology of Lactic Acid Bacteria. Novel Applications; Mozzi, F., Raya, R., Vignolo, G., Eds.; Wiley-Blackwell: Ames, ID, USA, 2010; pp. 251–271. ISBN 978-0-8138-1583-1. [Google Scholar]
- Paramithiotis, S.; Drosinos, E.; Sofos, J.; Nychas, G. Fermentation: Microbiology and Biochemistry. In Handbook of Meat Processing, 1st ed.; Toldrá, F., Ed.; Blackwell Publishing Inc.: Iowa, IA, USA, 2010; pp. 379–398. ISBN 978-0-8138-2182-5. [Google Scholar]
- Alquilante, L.; Santarelli, S.; Silvestri, G.; Osimani, A.; Petruzzelli, A.; Clementi, F. The microbial ecology of a typical Italian salami during its natural fermentation. Int. J. Food Microbiol. 2007, 120, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Dolci, P.; Rantsiou, K.; Urso, R.; Cantoni, C.; Comi, G. Lactic acid bacteria ecology of three traditional fermented sausages produced in the North of Italy as determined by molecular methods. Meat Sci. 2009, 82, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Rimaux, T.; Rivière, A.; Illeghems, K.; Weckx, S.; De Vuyst, L.; Leroy, F. Expression of the arginine deiminase pathway genes in Lactobacillus sakei is strain dependent and is affected by the environmental pH. Appl. Environ. Microbiol. 2012, 78, 4874–4883. [Google Scholar] [CrossRef] [PubMed]
- McLeod, A.; Snipen, L.; Naterstad, K.; Axelsson, L. Global transcriptome response in Lactobacillus sakei during growth on ribose. BMC Microbiol. 2011, 1, 145. [Google Scholar] [CrossRef] [PubMed]
- Urso, R.; Rantsiou, K.; Cantoni, C.; Comi, G.; Cocolin, L. Technological characterization of a bacteriocin-producing Lactobacillus sakei and its use in fermented sausages production. Int. J. Food Microbiol. 2006, 110, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Aymerich, M.T.; Garriga, M.; Monfort, J.M.; Nes, I.; Hugas, M. Bacteriocin-producing lactobacilli in Spanish-style fermented sausages: Characterization of bacteriocins. Food Microbiol. 2000, 17, 33–45. [Google Scholar] [CrossRef]
- Talon, R.; Leroy, S.; Lebert, I. Microbial ecosystems of traditional fermented meat products: The importance of indigenous starters. Meat Sci. 2007, 77, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Drosinos, E.H.; Mataragas, M.; Xiraphi, N.; Moschonas, G.; Gaitis, F.; Metaxopoulos, J. Characterization of the microbial flora from a traditional Greek fermented sausage. Meat Sci. 2005, 69, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Fadda, S.; Vignolo, G.; Oliver, G. Tyramine degradation and tyramine/histamine production by lactic acid bacteria and Kocuria strains. Biotechnol. Lett. 2001, 23, 2015–2019. [Google Scholar] [CrossRef]
- Demeyer, D. Meat fermentation: Principles and applications. In Handbook of Food and Beverage Fermentation Technology; Hui, H., Meunier-Goddik, L., Hansen, A., Josephsen, J., Nip, W.-K., Stanfield, P., Toldrá, F., Eds.; Marcel Dekker: New York, NY, USA, 2004; pp. 353–368. ISBN 0-8247-4780-1. [Google Scholar]
- Drosinos, E.H.; Paramithiotis, S.; Kolovos, G.; Tsikouras, I.; Metaxopoulos, I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in Southern Greece. Food Microbiol. 2007, 24, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Taboada, N.; Van Nieuwenhove, C.; Alzogaray, S.L.; Medina, R. Influence of autochthonous cultures on fatty acid composition, esterase activity and sensory profile of Argentinean goat cheeses. J. Food Compos. Anal. 2015, 40, 86–94. [Google Scholar] [CrossRef]
- Spaziani, M.; Del Torre, M.; Stecchini, M. Changes of physicochemical, microbiological, and textural properties during ripening of Italian low-acid sausages. Proteolysis, sensory and volatile profiles. Meat Sci. 2009, 81, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Fadda, S.; Vildoza, M.J.; Vignolo, G. The acidogenic metabolism of Lactobacillus plantarum CRL 681 improves sarcoplasmic protein hydrolysis during meat fermentation. J. Muscle Foods 2010, 21, 545–556. [Google Scholar] [CrossRef]
- Sanz, Y.; Sentandreu, M.A.; Toldra, F. Role of muscle and bacterial exopeptidases in meat fermentation. In Research Advances in the Quality of Meat and Meat Products; Toldra, F., Ed.; Research Singpost: Trivandrum, India, 2002; pp. 143–155. ISBN 81-7736-125-2. [Google Scholar]
- Freiding, S.; Gutsche, K.A.; Ehrmann, M.A.; Vogel, R.F. Genetic screening of Lactobacillus sakei and Lactobacillus curvatus strains for their peptidolytic system and amino acid metabolism, and comparison of their volatilomes in a model system. Syst. Appl. Microbiol. 2011, 34, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Ammor, S.; Dufour, E.; Zagorec, M.; Chaillou, S.; Chevallier, I. Characterization and selection of Lactobacillus sakei strains isolated from traditional dry sausage for their potential use as starter cultures. Food Microbiol. 2005, 22, 529–538. [Google Scholar] [CrossRef]
- Parente, E.; Grieco, S.; Crudele, M.A. Phenotypic diversity of lactic acid bacteria isolated from fermented sausages produced in Basilicata (Southern Italy). J. Appl. Microbiol. 2001, 90, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, M.G.; Ricciardi, A.; Zotta, T.; Parente, E.; Salzano, G. Molecular and technological characterization of lactic acid bacteria from traditional fermented sausages of Basilicata region (Southern Italy). Meat Sci. 2008, 80, 1238–1248. [Google Scholar] [CrossRef] [PubMed]

| pH | aw | ||||
|---|---|---|---|---|---|
| Time (days) | Formulation | A | B | A | B |
| 0 | 6.14 ± 0.07 A | 6.07 ± 0.07 A | 0.99 ± 0.01 C | 0.99 ± 0.01 C | |
| 3 | 5.36 ± 0.07 B | 5.52 ± 0.07 C | 0.91 ± 0.01 A | 0.94 ± 0.01 B | |
| 15 | 5.10 ± 0.07 D | 5.20 ± 0.07 E | 0.82 ± 0.01 D | 0.88 ± 0.01 E | |
| Time (Days) | 0 | 3 | 15 | |||
|---|---|---|---|---|---|---|
| Formulation | A | B | A | B | A | B |
| L* | 67.23 ± 0.96 D | 71.38 ± 0.96 E | 61.91 ± 0.96 C | 68.52 ± 0.96 D | 51.86 ± 0.96 A | 56.22 ± 0.96 B |
| a* | 3.84 ± 0.25 A | 3.53 ± 0.25 A | 4.90 ± 0.25 B | 4.04 ± 0.25 A | 16.21 ± 0.25 D | 14.55 ± 0.25 C |
| b* | 14.51 ± 0.15 D | 16.42 ± 0.15 E | 12.67 ± 0.15 B | 13.75 ± 0.15 C | 8.86 ± 0.15 A | 8.69 ± 0.15 A |
| C* | 15.01 ± 0.36 C | 16.80 ± 0.36 D | 13.58 ± 0.36 A | 14.33 ± 0.57 B | 16.82 ± 0.36 E | 13.62 ± 0.36 D |
| H* | 75.21 ± 0.57 D | 77.88 ± 0.57 D | 68.45 ± 0.57 C | 73.64 ± 0.57 D | 28.18 ± 0.57 A | 34.60 ± 0.57 B |
| Time Formulation/Microorganisms | 0 | 3 | 15 | |||
|---|---|---|---|---|---|---|
| A | B | A | B | A | B | |
| LAB a | 3.37 ± 0.37 A | 3.48 ± 0.37 A | 7.53 ± 0.37 B | 7.58 ± 0.37 B | 7.06 ± 0.37 B | 6.96 ± 0.37 B |
| GCC a | 2.08 ± 0.19 A | 2.09 ± 0.19 A | 6.26 ± 0.19 B | 6.09 ± 0.19 B | 6.09 ± 0.19 B | 5.88 ± 0.19 B |
| TMA a | 5.61 ± 0.12 A | 5.37 ± 0.12 A | 5.46 ± 0.12 A | 6.60 ± 0.12 B | 7.29 ± 0.12 C | 7.24 ± 0.09 C |
| Colif. 30 °C b | 4 A | <3 B | 15 C | <3 B | <3 B | <3 B |
| Colif. 45 °C b | <3 A | <3 A | <3 A | <3 A | <3 A | <3 A |
| S. aureus a | 3.00 ± 0.07 A | 5.30 ± 0.07 B | <2 A | <2 A | <2 A | <2 A |
| H & L a | 2.29 ± 0.26 A | 3.12 ± 0.26 B | 2.40 ± 0.26 AB | 4.17 ± 0.26 C | 2.47 ± 0.26 AB | 2.57 ± 0.26 AB |
| Salmonella/25 g a | Absence | Absence | Absence | Absence | Absence | Absence |
| Phenotypic Characterization | ||||||||
|---|---|---|---|---|---|---|---|---|
| Preliminary Identification/Phenotypic Tests | G1 L. sa | G2 L. cu | G3 L. pla | G4 L. catol | G5 L. carha | G6 L. ali | G7 L. bre | G8 L. far |
| Number of isolates | 41 a | 40 | 19 | 14 | 8 | 10 | 10 | 8 |
| Morphology | B | B | B | B | B | B | B | B |
| catalase activity | - | - | - | - | - | - | - | - |
| CO2 from glucose | - | - | - | - | - | - | + | - |
| CO2 from gluconate | + | + | + | 13 b | + | + | + | - |
| NH3 from arginine | + | - | - | - | - | + | - | + |
| Esculin hydrolysis | 61 | 80 | - | 93 | 87 | + | + | + |
| Growth with 4% NaCl | + | + | + | + | + | + | + | + |
| Growth with 10% NaCl | - | - | - | - | - | - | - | 90 |
| Growth at 15°C | + | + | + | + | + | + | + | + |
| Growth at 45°C | - | - | - | - | - | - | - | 90 |
| Acid production from: | ||||||||
| D (+)Fructose | + | + | + | + | + | + | + | + |
| L ramnose | - | - | - | - | + | - | - | - |
| Sucrose | + | 80 | 90 | - | + | + | + | + |
| Melezitose | - | - | 90 | - | 87 | - | - | - |
| Raffinose | - | - | 60 | - | - | - | + | - |
| Lactic acid production from: | ||||||||
| D (−) Arabinose | - | - | 69 | - | - | - | + | - |
| D (−) Lactose | 19 | 10 | 69 | - | 90 | + | + | - |
| D (−) Galactose | + | + | 74 | + | + | - | + | + |
| D (−) Mannose | 90 | + | 80 | + | + | 90 | 80 | + |
| D (−) Mannitol | - | - | 84 | - | + | + | - | + |
| D (−) Maltose | 93 | - | + | - | + | - | - | - |
| D (−) Ribose | 90 | + | 90 | 7 | + | + | + | + |
| D (−) Melibiose | + | - | 95 | - | 12 | - | 90 | - |
| D (−) Salicin | + | + | 68 | 14 | + | + | + | - |
| D (−) Glucose | + | + | + | - | + | + | - | + |
| Time Strains | 0 h | 24 h | 48 h | 72 h | 96 h | Net Change |
|---|---|---|---|---|---|---|
| Control * | 0.261 ** ± 0.02 | 0.266 ± 0.02 | 0.257 ± 0.02 | 0.257 ± 0.03 | 0.260 ± 0.06 | −0.001 |
| UL4 | 0.263 ± 0.01 | 0.287 ± 0.01 | 0.287 ± 0.04 | 0.283 ± 0.04 | 0.284 ± 0.05 | 0.021 |
| UL8 | 0.261 ± 0.03 | 0.266 ± 0.01 | 0.263 ± 0.01 | 0.279 ± 0.01 | 0.268 ± 0.02 | 0.007 |
| UL9 | 0.262 ± 0.01 | 0.278 ± 0.03 | 0.281 ± 0.03 | 0.352 ± 0.03 | 0.264 ± 0.04 | 0.002 |
| UL10 | 0.262 ± 0.02 | 0.277 ± 0.02 | 0.281 ± 0.03 | 0.328 ± 0.03 | 0.572 ± 0.07 | 0.300 |
| UL12 | 0.262 ± 0.01 | 0.372 ± 0.02 | 0.425 ± 0.02 | 0.360 ± 0.01 | 0.365 ± 0.01 | 0.103 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nediani, M.T.; García, L.; Saavedra, L.; Martínez, S.; López Alzogaray, S.; Fadda, S. Adding Value to Goat Meat: Biochemical and Technological Characterization of Autochthonous Lactic Acid Bacteria to Achieve High-Quality Fermented Sausages. Microorganisms 2017, 5, 26. https://doi.org/10.3390/microorganisms5020026
Nediani MT, García L, Saavedra L, Martínez S, López Alzogaray S, Fadda S. Adding Value to Goat Meat: Biochemical and Technological Characterization of Autochthonous Lactic Acid Bacteria to Achieve High-Quality Fermented Sausages. Microorganisms. 2017; 5(2):26. https://doi.org/10.3390/microorganisms5020026
Chicago/Turabian StyleNediani, Miriam T., Luis García, Lucila Saavedra, Sandra Martínez, Soledad López Alzogaray, and Silvina Fadda. 2017. "Adding Value to Goat Meat: Biochemical and Technological Characterization of Autochthonous Lactic Acid Bacteria to Achieve High-Quality Fermented Sausages" Microorganisms 5, no. 2: 26. https://doi.org/10.3390/microorganisms5020026