Purine Acquisition and Synthesis by Human Fungal Pathogens
Abstract
:1. The Diversity of Fungi and the Environments They Inhabit
2. Purines and Their Role in the Cell
3. Purines as a Nitrogen Source
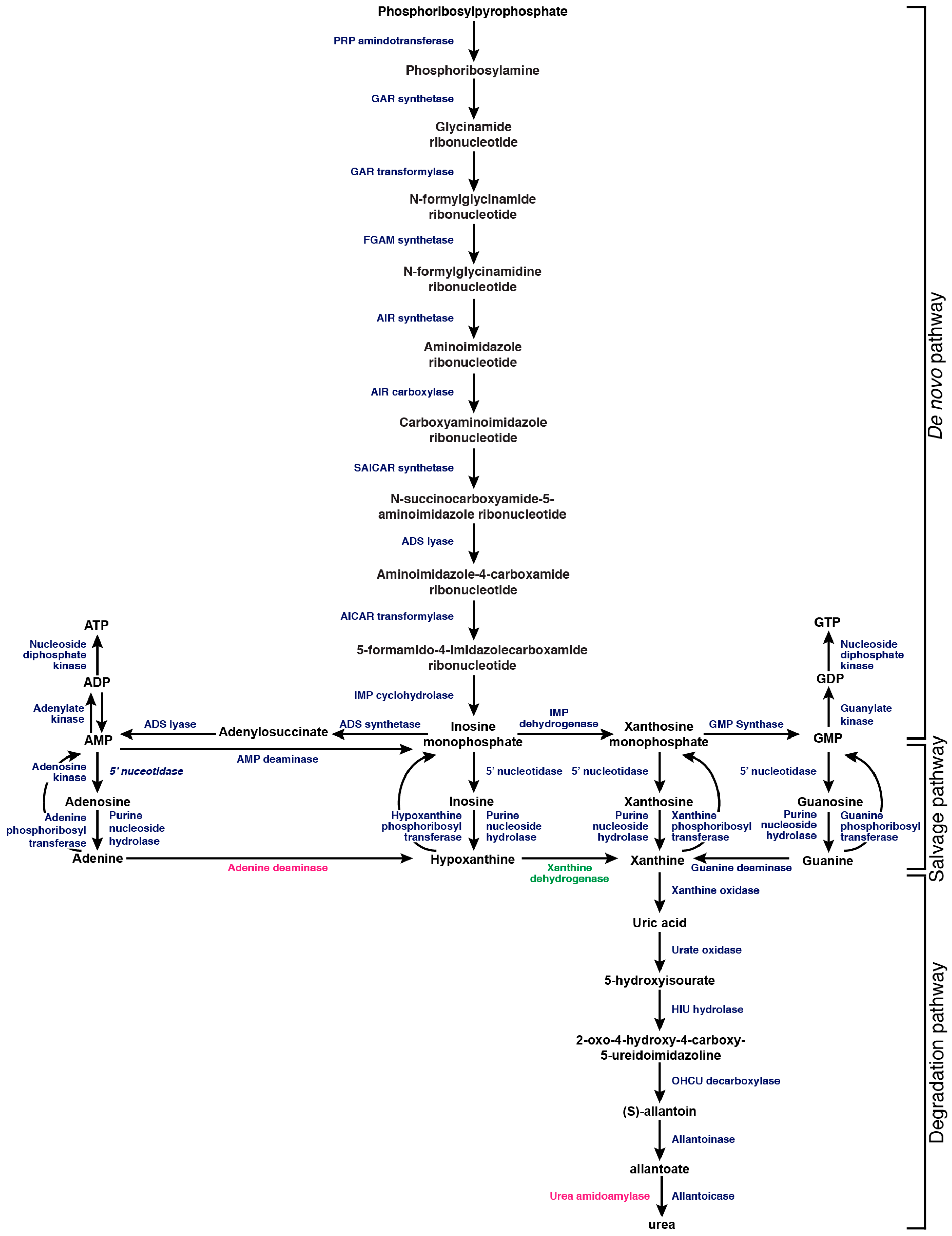
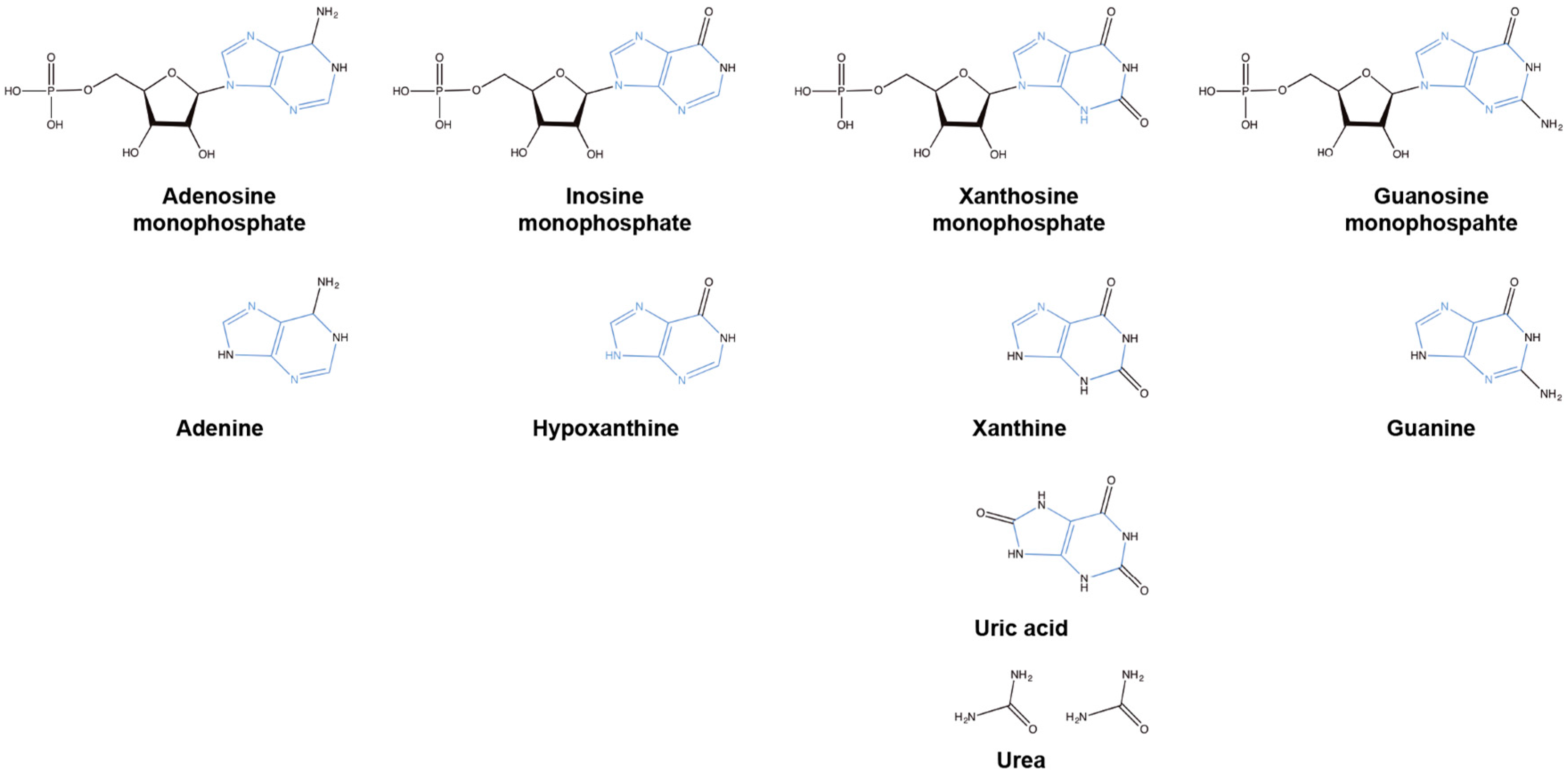
4. Salvaging Purines
5. Synthesizing Purines
6. Purine Metabolism in Candida Albicans
7. Purine Metabolism in Aspergillus Fumigatus
8. Purine Metabolism in Cryptococcus Neoformans
9. Purine Biosynthesis as an Antifungal Drug Target
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Clayton, M. Three newly published albums of seventeenth-century mycological drawings. Mycologist 2006, 20, 163–169. [Google Scholar] [CrossRef]
- Micheli, P.A. Nova Plantarum Genera. Available online: https://books.google.co.uk/books?hl=en&lr=&id=hIozjzXbJQEC&oi=fnd&pg=PA1&dq=Nova+Plantarum+Genera&ots=5W-BBlkw6g&sig=3SM31sAcacKOc5MUJzkwt1zbfcY#v=onepage&q=Nova%20Plantarum%20Genera&f=false (assessed on 7 June 2017).
- Linné, C.V. Systema Naturæ Per Regna Tria Naturæ, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis, 10th ed.; Impensis Direct; Laurentii Salvii: Holmiæ, Sweden, 1758. [Google Scholar]
- Persoon, C.H.; Lünemann, G.H. Synopsis Methodica Fungorum; Apud H. Dieterich: Gottingae, Germany, 1801. [Google Scholar]
- Fries, E.M. Systema Mycologicum, Sistens Fungorum Ordines, Genera et Species, huc Usque Cognitas; Ex Officina Berlingiana: Lundæ, Sweden, 1821. [Google Scholar]
- Petersen, R.H.; Knudsen, H. The mycological legacy of Elias Magnus Fries. IMA Fungus 2015, 6, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, R.H. New concepts of kingdoms or organisms. Evolutionary relations are better represented by new classifications than by the traditional two kingdoms. Science 1969, 163, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, D.L. Recent changes in the international rules affecting the nomenclature of fungi. Microbiol. Sci. 1984, 1, 18–21. [Google Scholar] [PubMed]
- Hawksworth, D.L. The fungal dimension of biodiversity—Magnitude, significance, and conservation. Mycol. Res. 1991, 95, 641–655. [Google Scholar] [CrossRef]
- Blackwell, M. The fungi: 1, 2, 3... 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Burgaud, G.; Le Calvez, T.; Arzur, D.; Vandenkoornhuyse, P.; Barbier, G. Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ. Microbiol. 2009, 11, 1588–1600. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, V.N.; Cantrell, C.L.; Wedge, D.E.; Ferreira, M.C.; Soares, M.A.; Jacob, M.R.; Oliveira, F.S.; Galante, D.; Rodrigues, F.; Alves, T.M.; et al. Fungi associated with rocks of the atacama desert: Taxonomy, distribution, diversity, ecology and bioprospection for bioactive compounds. Environ. Microbiol. 2016, 18, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Lyakh, S.P.; Kozlova, T.M.; Salivonik, S.M. Effect of periodic freezing and thawing on cells of the Antarctic black yeast nadsoniella-nigra var hesuelica. Microbiology 1983, 52, 486–491. [Google Scholar]
- Badiee, P.; Hashemizadeh, Z. Opportunistic invasive fungal infections: Diagnosis & clinical management. Indian J. Med. Res. 2014, 139, 195–204. [Google Scholar] [PubMed]
- Drgona, L.; Khachatryan, A.; Stephens, J.; Charbonneau, C.; Kantecki, M.; Haider, S.; Barnes, R. Clinical and economic burden of invasive fungal diseases in Europe: Focus on pre-emptive and empirical treatment of Aspergillus and Candida species. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Zilberberg, M.D.; Shorr, A.F.; Kollef, M.H. Secular trends in candidemia-related hospitalization in the United States, 2000–2005. Infect. Control Hosp. Epidemiol. 2008, 29, 978–980. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, R.P.; Gennings, C. Bloodstream infections due to Candida species in the intensive care unit: Identifying especially high-risk patients to determine prevention strategies. Clin. Infect. Dis. 2005, 41, 1694. [Google Scholar] [CrossRef] [PubMed]
- Kosmidis, C.; Denning, D.W. The clinical spectrum of pulmonary aspergillosis. Thorax 2015, 70, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R.; Dismukes, W.E.; Dromer, F.; Goldman, D.L.; Graybill, J.R.; Hamill, R.J.; Harrison, T.S.; Larsen, R.A.; Lortholary, O.; Nguyen, M.H.; et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of America. Clin. Infect. Dis. 2010, 50, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Rénon, L. Étude Sur L'aspergillose Chez Les Animaux Et Chez L’homme; Masson: Paris, France, 1897. [Google Scholar]
- Staib, F. Vogelkot, ein nahrsubstrat fur die gattung cryptococcus. Zentralbl. Bakteriol. 1962, 186, 233. [Google Scholar]
- Staib, F.; Seeliger, H.P. A new selective medium for the isolation of C. neoformans from fecal material and from soil. Ann. Inst. Pasteur 1966, 110, 792–793. [Google Scholar]
- Emmons, C.W. Saprophytic sources of Cryptococcus neoformans associated with the pigeon (Columba livia). Am. J. Hyg. 1955, 62, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Badiee, P.; Kordbacheh, P.; Alborzi, A.; Zeini, F.; Mirhendy, H.; Mahmoody, M. Fungal infections in solid organ recipients. Exp. Clin. Transpl. 2005, 3, 385–389. [Google Scholar]
- Nucci, M.; Anaissie, E. Revisiting the source of candidemia: Skin or gut? Clin. Infect. Dis. 2001, 33, 1959–1967. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.J. Integration of organ systems in avian osmoregulation. J. Exp. Zool. 1999, 283, 702–707. [Google Scholar] [CrossRef]
- Yamaoka, N.; Kaneko, K.; Kudo, Y.; Aoki, M.; Yasuda, M.; Mawatari, K.; Nakagomi, K.; Yamada, Y.; Yamamoto, T. Analysis of purine in purine-rich cauliflower. Nucleosides Nucleotides Nucleic Acids 2010, 29, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, O.; Shorey, E.C. Pyrimidine derivatives and purine bases in soils. J. Biol. Chem. 1910, 8, 385–393. [Google Scholar]
- Rodriguez-Nunez, A.; Camina, F.; Lojo, S.; Rodriguez-Segade, S.; Castro-Gago, M. Concentrations of nucleotides, nucleosides, purine bases and urate in cerebrospinal fluid of children with meningitis. Acta Paediatr. 1993, 82, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Fairbanks, L.D.; Harris, J.C.; Duley, J.A.; Simmonds, H.A. Nucleotide degradation products in cerebrospinal fluid (CSF) in inherited and acquired pathologies. Nucleosides Nucleotides Nucleic Acids 2004, 23, 1185–1187. [Google Scholar] [CrossRef] [PubMed]
- Eells, J.T.; Spector, R. Purine and pyrimidine base and nucleoside concentrations in human cerebrospinal fluid and plasma. Neurochem. Res. 1983, 8, 1451–1457. [Google Scholar] [CrossRef] [PubMed]
- Fischer, E.; Ach, L. Neue synthese der harnsäure und ihrer methylderivate. In Untersuchungen in der Puringruppe; Springer: Berlin, Germany, 1985. [Google Scholar]
- Rosemeyer, H. The chemodiversity of purine as a constituent of natural products. Chem. Biodivers. 2004, 1, 361–401. [Google Scholar] [CrossRef] [PubMed]
- Oparin, A.I.; Morgulis, S. The Origin of Life; The Macmillan Company: New York, NY, USA, 1938; p. 270. [Google Scholar]
- Miller, S.L.; Urey, H.C. Organic compound synthesis on the primitive earth. Science 1959, 130, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Marzluf, G.A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 1997, 61, 17–32. [Google Scholar] [PubMed]
- Lee, I.R.; Chow, E.W.; Morrow, C.A.; Djordjevic, J.T.; Fraser, J.A. Nitrogen metabolite repression of metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Genetics 2011, 188, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.H. Fungal Physiology, 2nd ed.; Wiley-Liss: New York, NY, USA, 1994; p. 458. [Google Scholar]
- Vogels, G.D.; Van der Drift, C. Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 1976, 40, 403–468. [Google Scholar] [PubMed]
- Sumrada, R.; Cooper, T.G. Allantoin transport in Saccharomyces cerevisiae. J. Bacteriol. 1977, 131, 839–847. [Google Scholar] [PubMed]
- Spanu, P.D. The genomics of obligate (and nonobligate) biotrophs. Annu. Rev. Phytopathol. 2012, 50, 91–109. [Google Scholar] [CrossRef] [PubMed]
- Cisse, O.H.; Pagni, M.; Hauser, P.M. Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol. Evol. 2014, 6, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [PubMed]
- Pantazopoulou, A.; Diallinas, G. Fungal nucleobase transporters. FEMS Microbiol. Rev. 2007, 31, 657–675. [Google Scholar] [CrossRef] [PubMed]
- Stouthamer, A.H. A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie van Leeuwenhoek 1973, 39, 545–565. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.; Milo, R. A feeling for the numbers in biology. Proc. Natl. Acad. Sci. USA 2009, 106, 21465–21471. [Google Scholar] [CrossRef] [PubMed]
- Dean, P.; Hirt, R.P.; Embley, T.M. Microsporidia: Why make nucleotides if you can steal them? PLoS Pathog. 2016, 12, e1005870. [Google Scholar] [CrossRef] [PubMed]
- Hirt, R.P.; Logsdon, J.M., Jr.; Healy, B.; Dorey, M.W.; Doolittle, W.F.; Embley, T.M. Microsporidia are related to fungi: Evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Galenus. Commentary on Hippocrates’ Epidemics Book I, Parts I-III; De Gruyter Akademie Forschung: Berlin, Germany, 2014; p. 736. [Google Scholar]
- Galen. Commentary on Hippocrates’ Epidemics Book II. Parts I-VI; Vagelpohl, U., Swain, S., Eds.; De Gruyter Akademie Forschung: Berlin, Germany, 2016; Volume 2, p. 1143. [Google Scholar]
- Knoke, M.; Bernhardt, H. The first description of an oesophageal candidosis by Bernhard von Langenbeck in 1839. Mycoses 2006, 49, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.A. A history of research on yeasts 8: Taxonomy. Yeast 2004, 21, 1141–1193. [Google Scholar] [CrossRef] [PubMed]
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldavs, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J.; et al. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014, 370, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Buffo, J.; Herman, M.A.; Soll, D.R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia 1984, 85, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, C.; Wang, A.; Liu, H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011, 9, e1001105. [Google Scholar] [CrossRef]
- Klengel, T.; Liang, W.J.; Chaloupka, J.; Ruoff, C.; Schroppel, K.; Naglik, J.R.; Eckert, S.E.; Mogensen, E.G.; Haynes, K.; Tuite, M.F.; et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 2005, 15, 2021–2026. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, C.; Liu, H. Candida albicans hyphal initiation and elongation. Trends Microbiol. 2014, 22, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K.; Aoyagi, Y.; Fukuuchi, T.; Inazawa, K.; Yamaoka, N. Total purine and purine base content of common foodstuffs for facilitating nutritional therapy for gout and hyperuricemia. Biol. Pharm. Bull. 2014, 37, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Karasawa, Y.; Ishi, I.T.; Kubota, T. Absorption and metabolism of purines by the small intestine of the chicken. Comp. Biochem. Physiol. A Comp. Physiol. 1991, 99, 235–240. [Google Scholar] [PubMed]
- Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, M.; Liang, X.; Jiang, Z.; Guo, X. Simultaneous determination of three purines in Alysicarpus vaginalis (l.) dc. By hollow fiber-based liquid-phase microextraction combined with high-performance liquid chromatography. Biomed. Chromatogr. 2014, 28, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Duan, J.A.; Qian, D.; Wang, H.; Tang, Y.; Qian, Y.; Wu, D.; Su, S.; Shang, E. Hydrophilic interaction ultra-high performance liquid chromatography coupled with triple quadrupole mass spectrometry for determination of nucleotides, nucleosides and nucleobases in Ziziphus plants. J. Chromatogr. A 2013, 1301, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Cortez, J.; Schnitzer, M. Purines and pyrimidines in soils and humic substances. Soil Sci. Soc. Am. J. 1979, 43, 958–961. [Google Scholar] [CrossRef]
- Tebung, W.A.; Choudhury, B.I.; Tebbji, F.; Morschhauser, J.; Whiteway, M. Rewiring of the ppr1 zinc cluster transcription factor from purine catabolism to pyrimidine biogenesis in the Saccharomycetaceae. Curr. Biol. 2016, 26, 1677–1687. [Google Scholar] [CrossRef] [PubMed]
- Vylkova, S.; Lorenz, M.C. Phagosomal neutralization by the fungal pathogen Candida albicans induces macrophage pyroptosis. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [PubMed]
- Balish, E.; Svihla, G. Ultraviolet microscopy of purines and amino acids in the vacuole of Candida albicans. J. Bacteriol. 1968, 96, 259–265. [Google Scholar] [PubMed]
- Rodrigues, L.; Russo-Abrahao, T.; Cunha, R.A.; Goncalves, T.; Meyer-Fernandes, J.R. Characterization of extracellular nucleotide metabolism in Candida albicans. FEMS Microbiol. Lett. 2016, 363, fnv212. [Google Scholar] [CrossRef] [PubMed]
- Poulter, R.T.; Rikkerink, E.H. Genetic analysis of red, adenine-requiring mutants of Candida albicans. J. Bacteriol. 1983, 156, 1066–1077. [Google Scholar] [PubMed]
- Donovan, M.; Schumuke, J.J.; Fonzi, W.A.; Bonar, S.L.; Gheesling-Mullis, K.; Jacob, G.S.; Davisson, V.J.; Dotson, S.B. Virulence of a phosphoribosylaminoimidazole carboxylase-deficient Candida albicans strain in an immunosuppressed murine model of systemic candidiasis. Infect. Immun. 2001, 69, 2542–2548. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Suarez, R.; Xu, D.; Veillette, K.; Davison, J.; Sillaots, S.; Kauffman, S.; Hu, W.; Bowman, J.; Martel, N.; Trosok, S.; et al. Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus. Chem. Biol. 2007, 14, 1163–1175. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhao, J.; Guo, R.; Li, J.; Yu, L.; Xu, D. Functional characterization and virulence study of ADE8 and GUA1 genes involved in the de novo purine biosynthesis in Candida albicans. FEMS Yeast Res. 2010, 10, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.; Burkard, L.; Panepinto, J.C. Inhibition of nucleotide biosynthesis potentiates the antifungal activity of amphotericin b. PLoS ONE 2014, 9, e87246. [Google Scholar] [CrossRef] [PubMed]
- Plaignaud, M. Observation sur un fungus du sinus maxillaire. J. Méd. Chir. Pharm. 1791, 87, 244–251. [Google Scholar]
- Schmidt, A.; Schmidt, D.I. J.B. Georg W. Fresenius and the description of the species Aspergillus fumigatus in 1863. Contrib. Microbiol. 1999, 2, 1–4. [Google Scholar] [PubMed]
- Ruchlemer, R.; Yinnon, A.M.; Hershko, C. Changes in the natural history of invasive pulmonary aspergillosis in neutropenic leukemic patients. Isr. J. Med. Sci. 1996, 32, 1089–1092. [Google Scholar] [PubMed]
- Cohen, J.; Denning, D.W.; Viviani, M.A. Epidemiology of invasive aspergillosis in European cancer centres. Eortc invasive fungal infections cooperative group. Eur. J. Clin. Microbiol. Infect. Dis. 1993, 12, 392–393. [Google Scholar] [CrossRef] [PubMed]
- Groll, A.H.; Shah, P.M.; Mentzel, C.; Schneider, M.; Just-Nuebling, G.; Huebner, K. Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J. Infect. 1996, 33, 23–32. [Google Scholar] [CrossRef]
- Latge, J.P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999, 12, 310–350. [Google Scholar] [PubMed]
- Bodey, G.; Bueltmann, B.; Duguid, W.; Gibbs, D.; Hanak, H.; Hotchi, M.; Mall, G.; Martino, P.; Meunier, F.; Milliken, S.; et al. Fungal infections in cancer patients: An international autopsy survey. Eur. J. Clin. Microbiol. Infect. Dis. 1992, 11, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Elion, G.B. An overview of the role of nucleosides in chemotherapy. Adv. Enzyme Regul. 1985, 24, 323–334. [Google Scholar] [CrossRef]
- Patel, R.; Paya, C.V. Infections in solid-organ transplant recipients. Clin. Microbiol. Rev. 1997, 10, 86–124. [Google Scholar] [PubMed]
- Denning, D.W. Issues in the management of invasive aspergillosis. Ann. Med. Interne 1995, 146, 106–110. [Google Scholar]
- Denning, D.W. Therapeutic outcome in invasive aspergillosis. Clin. Infect. Dis. 1996, 23, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, T.R.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 2009, 22, 447–465. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Sugui, J.A. Aspergillus fumigatus—What makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013, 9, e1003743. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, H.; Amaya, F.; Hashimoto, S.; Ueno, H.; Beppu, S.; Mizuta, M.; Shime, N.; Ishizaka, A.; Hashimoto, S. Acute lung inflammation and ventilator-induced lung injury caused by ATP via the p2y receptors: An experimental study. Respir. Res. 2008, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Gournas, C.; Oestreicher, N.; Amillis, S.; Diallinas, G.; Scazzocchio, C. Completing the purine utilisation pathway of Aspergillus nidulans. Fungal Genet. Biol. 2011, 48, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Scazzocchio, C. The purine degradation pathway, genetics, biochemistry and regulation. Prog. Ind. Microbiol. 1994, 29, 221–257. [Google Scholar] [PubMed]
- Scazzocchio, C.; Darlington, A.J. The genetic control of xanthine dehydrogenase and urate oxidase synthess in Aspergillus nidulans. Bull. Soc. Chim. Biol. 1967, 49, 1503–1508. [Google Scholar] [PubMed]
- Scazzocchio, C.; Sdrin, N.; Ong, G. Positive regulation in a eukaryote, a study of the uaY gene of Aspergillus nidulans: I. Characterization of alleles, dominance and complementation studies, and a fine structure map of the uaY--oxpa cluster. Genetics 1982, 100, 185–208. [Google Scholar] [PubMed]
- Oestreicher, N.; Ribard, C.; Scazzocchio, C. The nada gene of Aspergillus nidulans, encoding adenine deaminase, is subject to a unique regulatory pattern. Fungal Genet. Biol. 2008, 45, 760–775. [Google Scholar] [CrossRef] [PubMed]
- Thykaer, J.; Andersen, M.R.; Baker, S.E. Essential pathway identification: From in silico analysis to potential antifungal targets in Aspergillus fumigatus. Med. Mycol. 2009, 47 (Suppl. 1), S80–S87. [Google Scholar] [CrossRef] [PubMed]
- Sanfelice, F. Sull’azione patogena dei bastomiceti [on the action of pathogenic bastomiceti]. Ann. Inst Igien. Univ. Roma 1895, 5, 239–262. [Google Scholar]
- Knoke, M.; Schwesinger, G. One hundred years ago: The history of cryptococcosis in greifswald. Medical mycology in the nineteenth century. Mycoses 1994, 37, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Dix, N.J.; Webster, J. Fungal Ecology; Chapman & Hall: London, UK, 1995; Volume VIII, p. 549. [Google Scholar]
- Baldrian, P.; Valaskova, V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 2008, 32, 501–521. [Google Scholar] [CrossRef] [PubMed]
- Stubblefield, S.P.; Taylor, T.N.; Beck, C.B. Studies of paleozoic fungi. 4. Wood-decaying fungi in callixylon-newberryi from the upper devonian. Am. J. Bot. 1985, 72, 1765–1774. [Google Scholar] [CrossRef]
- Rao, K.P.; Gopalakrishnareddy, T. Nitrogen excretion in arachnids. Comp. Biochem. Physiol. 1962, 7, 175–178. [Google Scholar] [CrossRef]
- Schmidt, G.; Liss, M.; Thannhauser, S.J. Guanine, the principal nitrogenous component of the excrements of certain spiders. Biochim. Biophys. Acta 1955, 16, 533–535. [Google Scholar] [CrossRef]
- Quilter, J. Moche politics, religion, and warfare. J. World Prehist. 2002, 16, 145–195. [Google Scholar] [CrossRef]
- International Union of American Republics. Bulletin of the International Union of the American Republics; International Bureau of the American Republics: Washington, DC, USA, 1909; p. 699. [Google Scholar]
- Magnus. Ueber das vorkommen von xanthicoxyd im guano. Ann. Chem. Pharm. 1844, 51, 395–397. [Google Scholar]
- Nielsen, K.; De Obaldia, A.L.; Heitman, J. Cryptococcus neoformans mates on pigeon guano: Implications for the realized ecological niche and globalization. Eukaryot. Cell 2007, 6, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.R.; Yang, L.; Sebetso, G.; Allen, R.; Doan, T.H.; Blundell, R.; Lui, E.Y.; Morrow, C.A.; Fraser, J.A. Characterization of the complete uric acid degradation pathway in the fungal pathogen Cryptococcus neoformans. PLoS ONE 2013, 8, e64292. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Li, S.S.; Zheng, C.; Jones, G.J.; Kim, K.S.; Zhou, H.; Kubes, P.; Mody, C.H. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J. Clin. Investig. 2010, 120, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, M.A.; Noverr, M.C.; Chen, G.H.; Toews, G.B.; Cox, G.M.; Perfect, J.R.; Huffnagle, G.B. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am. J. Pathol. 2004, 164, 1761–1771. [Google Scholar] [CrossRef]
- Cox, G.M.; Mukherjee, J.; Cole, G.T.; Casadevall, A.; Perfect, J.R. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 2000, 68, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Morrow, C.A.; Valkov, E.; Stamp, A.; Chow, E.W.; Lee, I.R.; Wronski, A.; Williams, S.J.; Hill, J.M.; Djordjevic, J.T.; Kappler, U.; et al. De novo GTP biosynthesis is critical for virulence of the fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2012, 8, e1002957. [Google Scholar] [CrossRef] [PubMed]
- Blundell, R.D.; Williams, S.J.; Arras, S.D.M.; Chitty, J.L.; Blake, K.L.; Ericsson, D.J.; Tibrewal, N.; Rohr, J.; Koh, Y.Q.A.E.; Kappler, U.; et al. Disruption of de novo adenosine triphosphate (ATP) biosynthesis abolishes virulence in Cryptococcus neoformans. ACS Infect. Dis. 2016, 2, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Arras, S.D.; Chitty, J.L.; Blake, K.L.; Schulz, B.L.; Fraser, J.A. A genomic safe haven for mutant complementation in Cryptococcus neoformans. PLoS ONE 2015, 10, e0122916. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R.; Toffaletti, D.L.; Rude, T.H. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun. 1993, 61, 4446–4451. [Google Scholar] [PubMed]
- Firestine, S.M.; Misialek, S.; Toffaletti, D.L.; Klem, T.J.; Perfect, J.R.; Davisson, V.J. Biochemical role of the Cryptococcus neoformans ADE2 protein in fungal de novo purine biosynthesis. Arch. Biochem. Biophys. 1998, 351, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Chitty, J.L.; Tatzenko, T.L.; Williams, S.J.; Koh, Y.Q.; Corfield, E.C.; Butler, M.S.; Robertson, A.A.; Cooper, M.A.; Kappler, U.; Kobe, B.; et al. GMP synthase is required for virulence factor production and infection by Cryptococcus neoformans. J. Biol. Chem. 2017, 292, 3049–3059. [Google Scholar] [CrossRef] [PubMed]
- Chitty, J.L.; Blake, K.L.; Blundell, R.D.; Koh, Y.Q.A.E.; Thompson, M.; Robertson, A.A.B.; Butler, M.S.; Cooper, M.A.; Kappler, U.; Williams, S.J.; et al. Cryptococcus neoformans ADS lyase in an enzyme essential for virulence whose crystal structure reveals features exploitable in antifungal drug design. J. Biol. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hitchings, G.H.; Elion, G.B.; Vanderwerff, H. 2-aminopurine as a purine antagonist. Fed. Proc. 1948, 7, 160. [Google Scholar] [PubMed]
- Elion, G.B. Nobel lecture. The purine path to chemotherapy. Biosci. Rep. 1989, 9, 509–529. [Google Scholar] [CrossRef] [PubMed]
- Shigeura, H.T.; Gordon, C.N. Hadacidin, a new inhibitor of purine biosynthesis. J. Biol. Chem. 1962, 237, 1932–1936. [Google Scholar] [PubMed]
- Christopherson, R.I.; Lyons, S.D.; Wilson, P.K. Inhibitors of de novo nucleotide biosynthesis as drugs. Acc. Chem. Res. 2002, 35, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Skipper, H.E.; Thomson, J.R.; Elion, G.B.; Hitchings, G.H. Observations on the anticancer activity of 6-mercaptopurine. Cancer Res. 1954, 14, 294–298. [Google Scholar] [PubMed]
- Mendelsohn, L.G.; Shih, C.; Schultz, R.M.; Worzalla, J.F. Biochemistry and pharmacology of glycinamide ribonucleotide formyltransferase inhibitors: Ly309887 and lometrexol. Investig. New Drugs 1996, 14, 287–294. [Google Scholar] [CrossRef]
- Franklin, T.J.; Cook, J.M. The inhibition of nucleic acid synthesis by mycophenolic acid. Biochem. J. 1969, 113, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.J.; Hoffman, D.H.; Esterman, M.A. Metabolism and biochemistry of mycophenolic acid. Cancer Res. 1972, 32, 1803–1809. [Google Scholar] [PubMed]
- Kohler, G.A.; Gong, X.; Bentink, S.; Theiss, S.; Pagani, G.M.; Agabian, N.; Hedstrom, L. The functional basis of mycophenolic acid resistance in Candida albicans IMP dehydrogenase. J. Biol. Chem. 2005, 280, 11295–11302. [Google Scholar] [CrossRef] [PubMed]
- Mezger, M.; Wozniok, I.; Blockhaus, C.; Kurzai, O.; Hebart, H.; Einsele, H.; Loeffler, J. Impact of mycophenolic acid on the functionality of human polymorphonuclear neutrophils and dendritic cells during interaction with Aspergillus fumigatus. Antimicrob. Agents Chemother. 2008, 52, 2644–2646. [Google Scholar] [CrossRef] [PubMed]
- Guillen Schlippe, Y.V.; Riera, T.V.; Seyedsayamdost, M.R.; Hedstrom, L. Substitution of the conserved arg-tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase reaction. Biochemistry 2004, 43, 4511–4521. [Google Scholar] [CrossRef] [PubMed]
- Schneweis, I.; Meyer, K.; Hormansdorfer, S.; Bauer, J. Mycophenolic acid in silage. Appl. Environ. Microbiol. 2000, 66, 3639–3641. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Mode of action of hadacidin in the growing bacterial cell. Nature 1966, 212, 93–94. [Google Scholar] [CrossRef] [PubMed]
- Tibrewal, N.; Elliott, G.I. Evaluation of hadacidin analogues. Bioorg. Med. Chem. Lett. 2011, 21, 517–519. [Google Scholar] [CrossRef] [PubMed]
| Source of purine | Adenine | Guanine | Xanthine | Hypoxanthine | Inosine | Reference |
|---|---|---|---|---|---|---|
| Average meal 1 (per gram) | 0.9 | 1.0 | 1.8 | 0.02 | ND | [58] |
| Human Blood serum | 0.4 | 97 | 20 | 172 | 168 | [60] |
| Human Cerebral spinal fluid | 0.2 | 0.5 | 2.4 | 3.9 | 0.6 | [29,30] |
| Human Intracellular | 1.5 | 97 | ND | 370 | 211 | [60] |
| Plant matter average 2 | 0.4 μg/mL | 1.3 μg/mL | 0.8 μg/mL | 1.0 μg/mL | 1.2 μg/mL | [61,62] |
| Soil average 3 | 19 M % | 19 M % | ND | ND | ND | [63] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chitty, J.L.; Fraser, J.A. Purine Acquisition and Synthesis by Human Fungal Pathogens. Microorganisms 2017, 5, 33. https://doi.org/10.3390/microorganisms5020033
Chitty JL, Fraser JA. Purine Acquisition and Synthesis by Human Fungal Pathogens. Microorganisms. 2017; 5(2):33. https://doi.org/10.3390/microorganisms5020033
Chicago/Turabian StyleChitty, Jessica L., and James A. Fraser. 2017. "Purine Acquisition and Synthesis by Human Fungal Pathogens" Microorganisms 5, no. 2: 33. https://doi.org/10.3390/microorganisms5020033






