Identification of sucA, Encoding β-Fructofuranosidase, in Rhizopus microsporus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Analyses of Organic Acids Production and Sugar Hydrolysis
2.3. Preparation of Cell-Free Extracts
2.4. Enzyme Assays
2.5. Identification and Sequencing of sucA Gene
3. Results
3.1. Detection of β-Fructofuranosidase (sucA) Gene and Production of Organic Acids and Ethanol in Rhizopus Strains
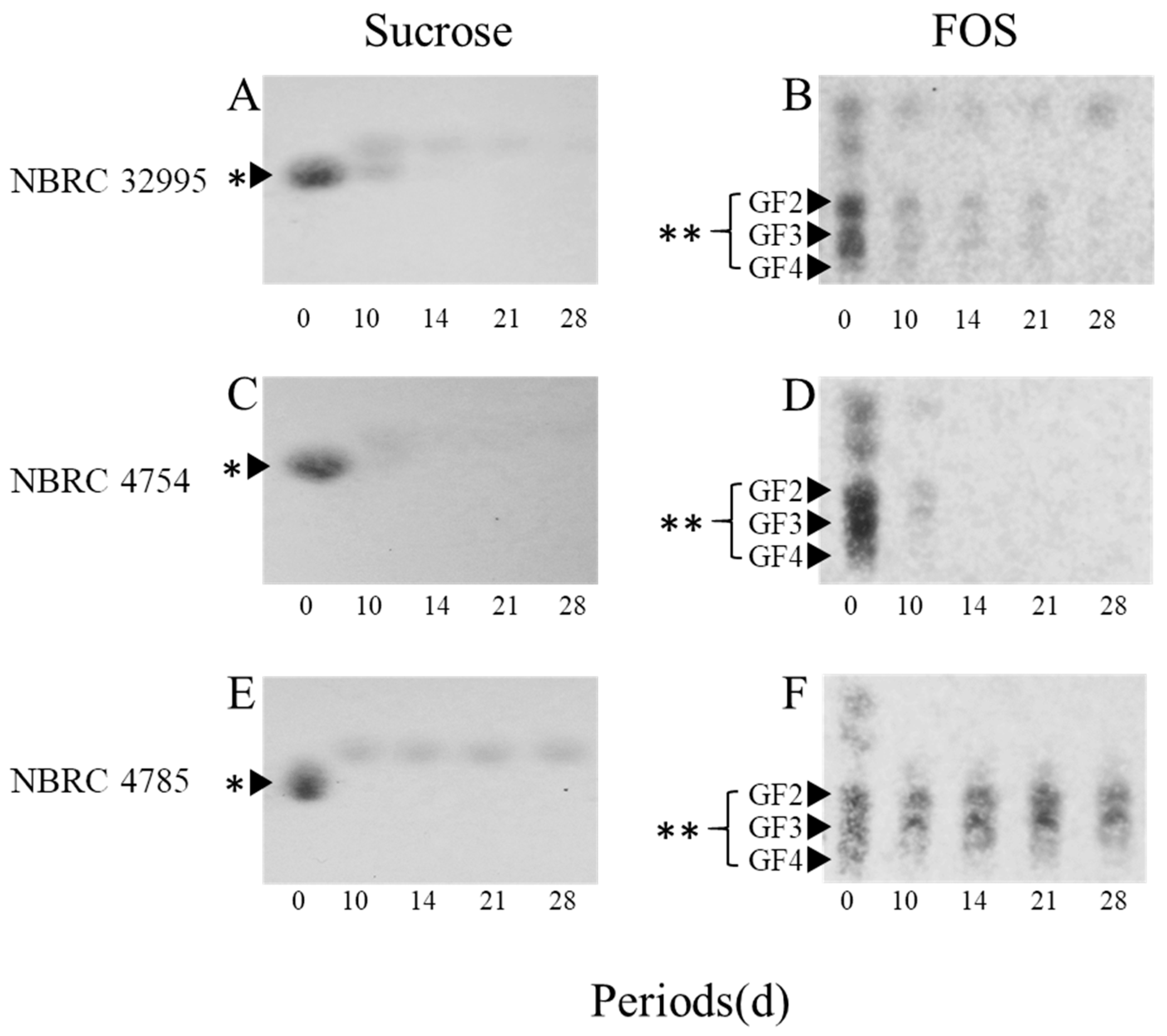
3.2. Analysis of Sucrose and FOS Hydrolysis by TLC
3.3. Sucrose-Hydrolyzing Activity and pH in the Culture Medium
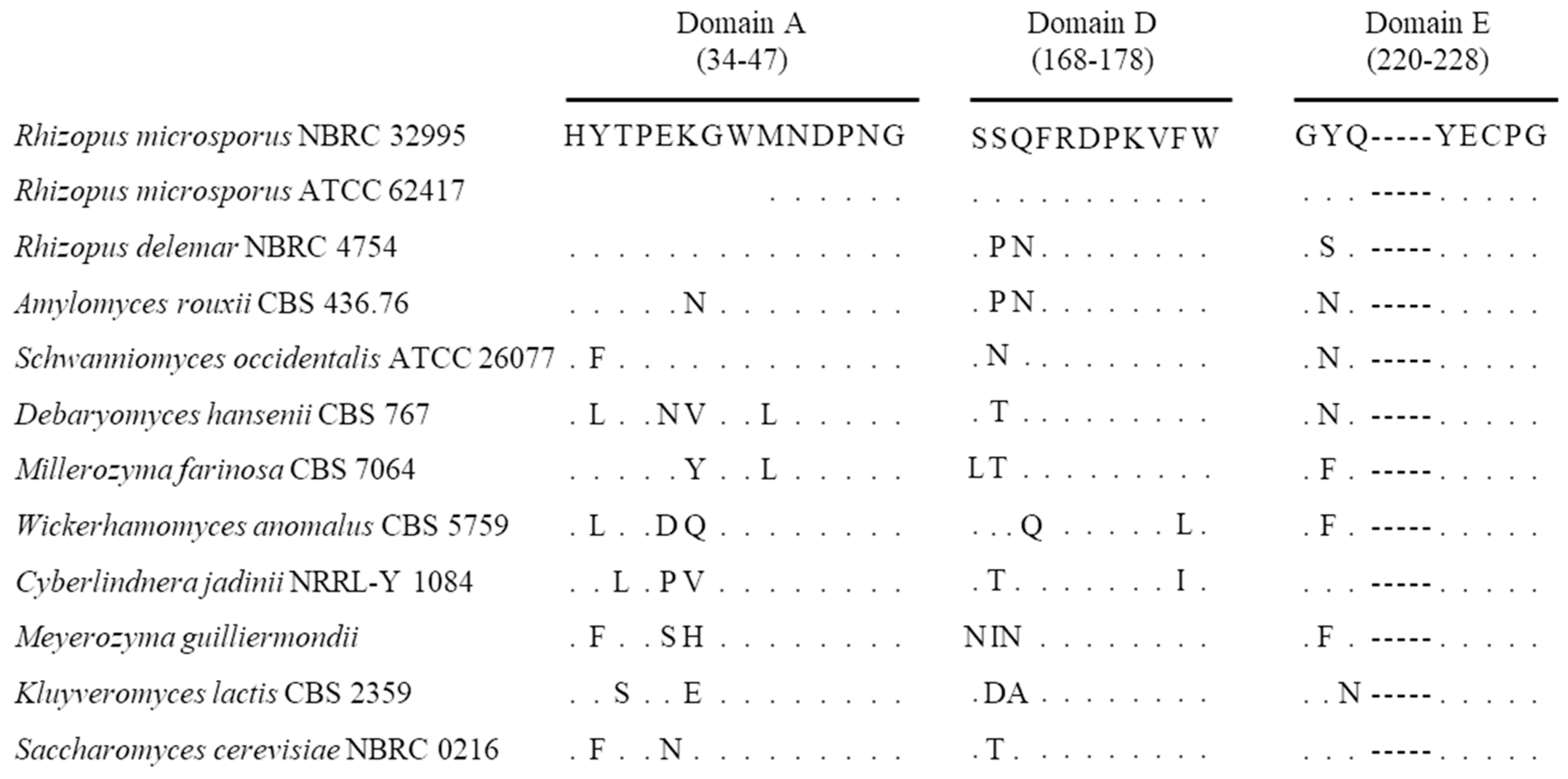
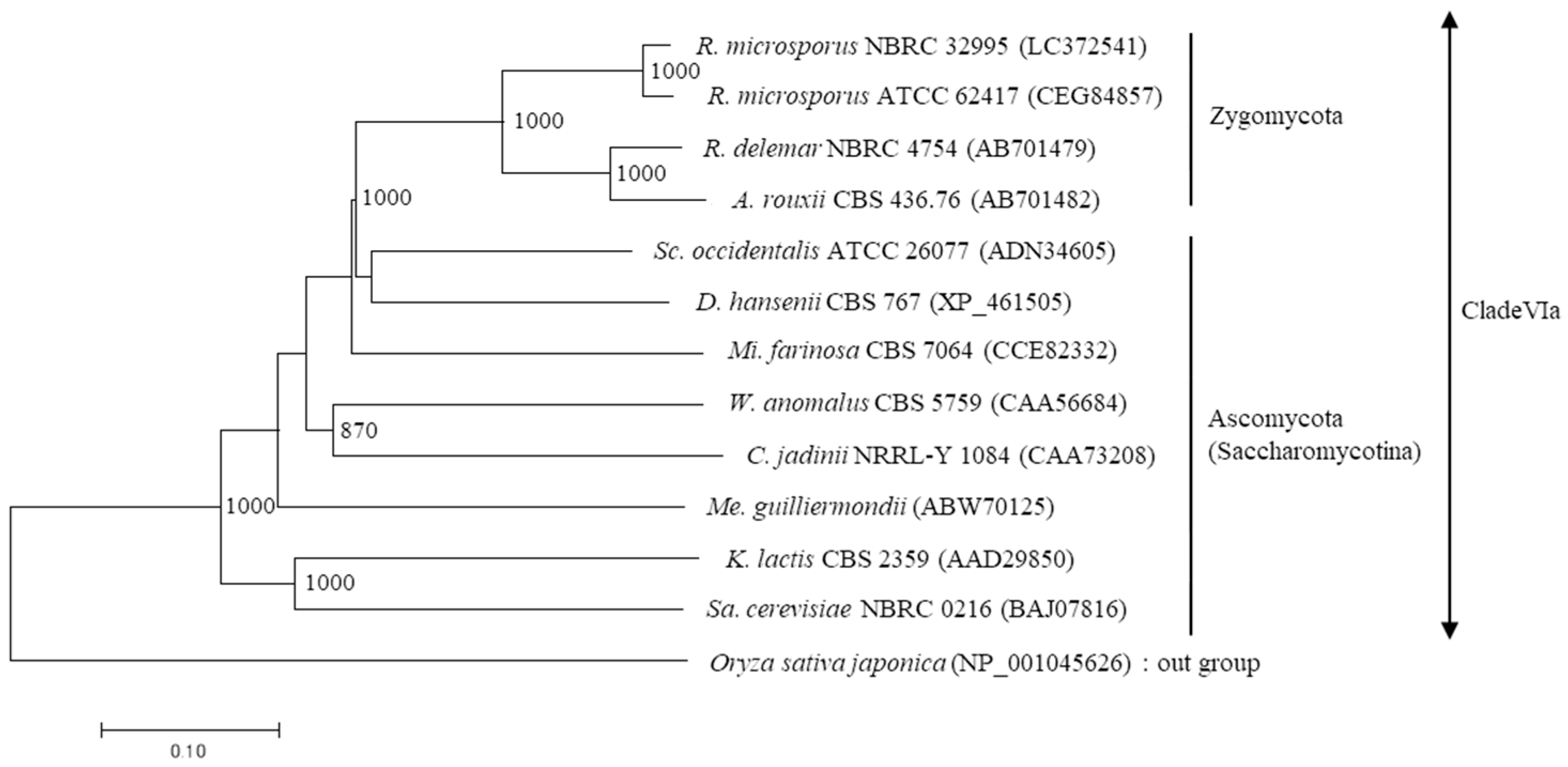
3.4. Sequencing and Phylogenetic Analysis of β-Fructofuranosidase (sucA) Gene
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Severo, C.B.; Guazzelli, L.S.; Severo, L.C. Chapter 7: Zygomycosis. J. Bras. Pneumol. 2010, 36, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Hesseltine, C.W. Microbiology of oriental fermented foods. Ann. Rev. Microbiol. 1983, 37, 575–601. [Google Scholar] [CrossRef] [PubMed]
- Rehms, H.; Barz, W. Degradation of stachyose, raffinose, melibiose and sucrose by different tempe-producing Rhizopus fungi. Appl. Microbiol. Biotechnol. 1995, 44, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Sone, T.; Sujaya, I.-N.; Saito, K.; Oda, Y.; Asano, K.; Tomita, T. rDNA ITS sequence of Rhizopus oryzae: Its application to classification and identification of lactic acid producers. Biosci. Biotechnol. Biochem. 2003, 67, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Oda, Y.; Asano, K.; Sone, T. Rhizopus delemar is the proper name for Rhizopus oryzae fumaric-malic acid producers. Mycologia 2007, 99, 714–722. [Google Scholar] [CrossRef] [PubMed]
- NU-IUBMB.; Webb, E.C. Enzyme Nomenclature; Academic Press: San Diego, CA, USA, 1992; pp. 346–369. ISBN 0-12-227164-5. [Google Scholar]
- Watanabe, T.; Oda, Y. Comparison of sucrose-hydrolyzing enzymes produced by Rhizopus oryzae and Amylomyces rouxii. Biosci. Biotechnol. Biochem. 2008, 72, 3167–3173. [Google Scholar] [CrossRef] [PubMed]
- Orikasa, Y.; Oda, Y. Molecular characterization of β-fructofuranosidases from Rhizopus delemar and Amylomyces rouxii. Folia Microbiol. 2013, 58, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Alméciga-Díaz, C.J.; Gutierrez, A.M.; Bahamon, I.; Rodríguez, A.; Rodríguez, M.A.; Sánchez, O.F. Computational analysis of the fructosyltransferase enzymes in plants, fungi and bacteria. Gene 2011, 484, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Schipper, M.A.A. A revision of the genus Rhizopus: I. The Rhizopus stolonifer-group and Rhizopus oryzae. Stud. Mycol. 1983, 25, 1–19. [Google Scholar]
- Schipper, M.A.A.; Stalpers, J.A. A revision of the genus Rhizopus: II. The Rhizopus microsporus group. Stud. Mycol. 1984, 25, 20–34. [Google Scholar]
- Kitpreechavanich, V.; Maneeboon, T.; Kayano, Y.; Sakai, K. Comparative characterization of l-lactic acid-producing thermotolerant Rhizopus fungi. J. Biosci. Bioeng. 2008, 106, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Ouchi, K. Construction of a sucrose-fermenting baker’s yeast incapable of hydrolyzing fructooligosaccharides. Enzyme Microb. Technol. 1991, 13, 495–498. [Google Scholar] [CrossRef]
- Oda, Y.; Tonomura, K. Purification and characterization of invertase from Torulaspora pretoriensis YK-1. Biosci. Biotechnol. Biochem. 1994, 58, 1155–1157. [Google Scholar] [CrossRef]
- Sone, T.; Abe, T.; Yoshida, N.; Suto, M.; Tomita, F. DNA fingerprinting and electrophoretic karyotyping of Japanese isolates of rice blast fungus. Ann. Phytopathol. Soc. Jpn. 1997, 63, 155–163. [Google Scholar] [CrossRef]
- Milne, I.; Bayer, M.; Cardle, L.; Shaw, P.; Stephen, G.; Wright, F.; Marshall, D. Tablet-next generation sequence assembly visualization. Bioinformatics 2009, 26, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The carbohydrate-active enzymes database (CAZy): An expert for glycogenomics. Nucl. Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Horn, F.; Guthke, R.; Hertweck, C. Draft genome sequences of symbiotic and nonsymbiotic Rhizopus microsporus strains CBS 344.29 and ATCC 62417. Genome Announc. 2015, 3, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Flores-Gallegos, A.C.; Contreras-Esquivel, J.C.; Morlett-Chávez, J.A.; Aguilar, C.N.; Rodríguez-Herrera, R. Comparative study of fungal strains for thermostable inulinase production. J. Biosci. Bioeng. 2015, 119, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Ohta, K.; Suetsugu, N.; Nakamura, T. Purification and properties of an extracellular inulinase from Rhizopus sp. strain TN-96. J. Biosci. Bioeng. 2002, 94, 78–80. [Google Scholar] [CrossRef]

| Strains | sucA 1 | Concentration (mg/mL) | References | ||||
|---|---|---|---|---|---|---|---|
| Malic Acid | Lactic Acid | Fumaric Acid | Ethanol | ||||
| Rhizopus oryzae | NBRC 4785 | − | 6.34 ± 1.53 | 93.67 ± 14.46 | 0.72 ± 0.45 | 5.80 ± 1.90 | Watanabe and Oda [7] |
| Rhizopus delemar | NBRC 4754 | + | 34.26 ± 10.03 | 8.18 ± 1.81 | 30.28 ± 9.74 | 30.71 ± 9.36 | Orikasa and Oda [8] |
| R. microsporus var. chinensis | NBRC 4737 | − | n.d. 2 | 0.25 ± 0.10 | n.d. | 0.24 ± 0.10 | This study |
| R. microsporus var. chinensis | NBRC 4768 | − | n.d. | 0.11 ± 0.07 | n.d. | 1.44 ± 0.56 | This study |
| R. microsporus var. chinensis | NBRC 31988 | − | n.d. | 1.00 ± 0.71 | n.d. | 1.91 ± 0.58 | This study |
| R. microsporus var. microsporus | NBRC 32995 | + | 32.71 ± 6.42 | 58.06 ± 6.36 | 7.87 ± 1.41 | 19.60 ± 0.66 | This study |
| R. microsporus var. microsporus | NBRC 32996 | − | 0.44 ± 0.11 | 0.31 ± 0.05 | n.d. | 0.41 ± 0.07 | This study |
| R. microsporus var. rhizopodiformis | NBRC 32997 | − | n.d. | n.d. | n.d. | 0.06 ± 0.01 | This study |
| R. microsporus var. tuberosus | NBRC 100014 | − | n.d. | 0.18 ± 0.07 | n.d. | 0.15 ± 0.03 | This study |
| R. stolonifer var. lyococcus | NBRC 32998 | − | 2.44 ± 0.58 | n.d. | n.d. | 0.14 ± 0.04 | This study |
| R. stolonifer var. stolonifer | NBRC 4781 | − | 9.24 ± 1.50 | 81.26 ± 10.41 | 5.63 ± 1.32 | 14.32 ± 1.74 | This study |
| R. stolonifer var. stolonifer | NBRC 5411 | − | 0.93 ± 0.41 | 0.25 ± 0.06 | n.d. | 0.23 ± 0.04 | This study |
| R. stolonifer var. stolonifer | NBRC 6188 | − | 1.32 ± 0.41 | 0.47 ± 0.22 | n.d. | 2.29 ± 0.39 | This study |
| R. stolonifer var. stolonifer | NBRC 30816 | − | 1.78 ± 0.34 | 0.26 ± 0.08 | n.d. | 1.36 ± 0.32 | This study |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orikasa, Y.; Oda, Y.; Ohwada, T. Identification of sucA, Encoding β-Fructofuranosidase, in Rhizopus microsporus. Microorganisms 2018, 6, 26. https://doi.org/10.3390/microorganisms6010026
Orikasa Y, Oda Y, Ohwada T. Identification of sucA, Encoding β-Fructofuranosidase, in Rhizopus microsporus. Microorganisms. 2018; 6(1):26. https://doi.org/10.3390/microorganisms6010026
Chicago/Turabian StyleOrikasa, Yoshitake, Yuji Oda, and Takuji Ohwada. 2018. "Identification of sucA, Encoding β-Fructofuranosidase, in Rhizopus microsporus" Microorganisms 6, no. 1: 26. https://doi.org/10.3390/microorganisms6010026




