Environmental Filtering Drives the Assembly of Habitat Generalists and Specialists in the Coastal Sand Microbial Communities of Southern China
Abstract
:1. Introduction
2. Materials and Methods
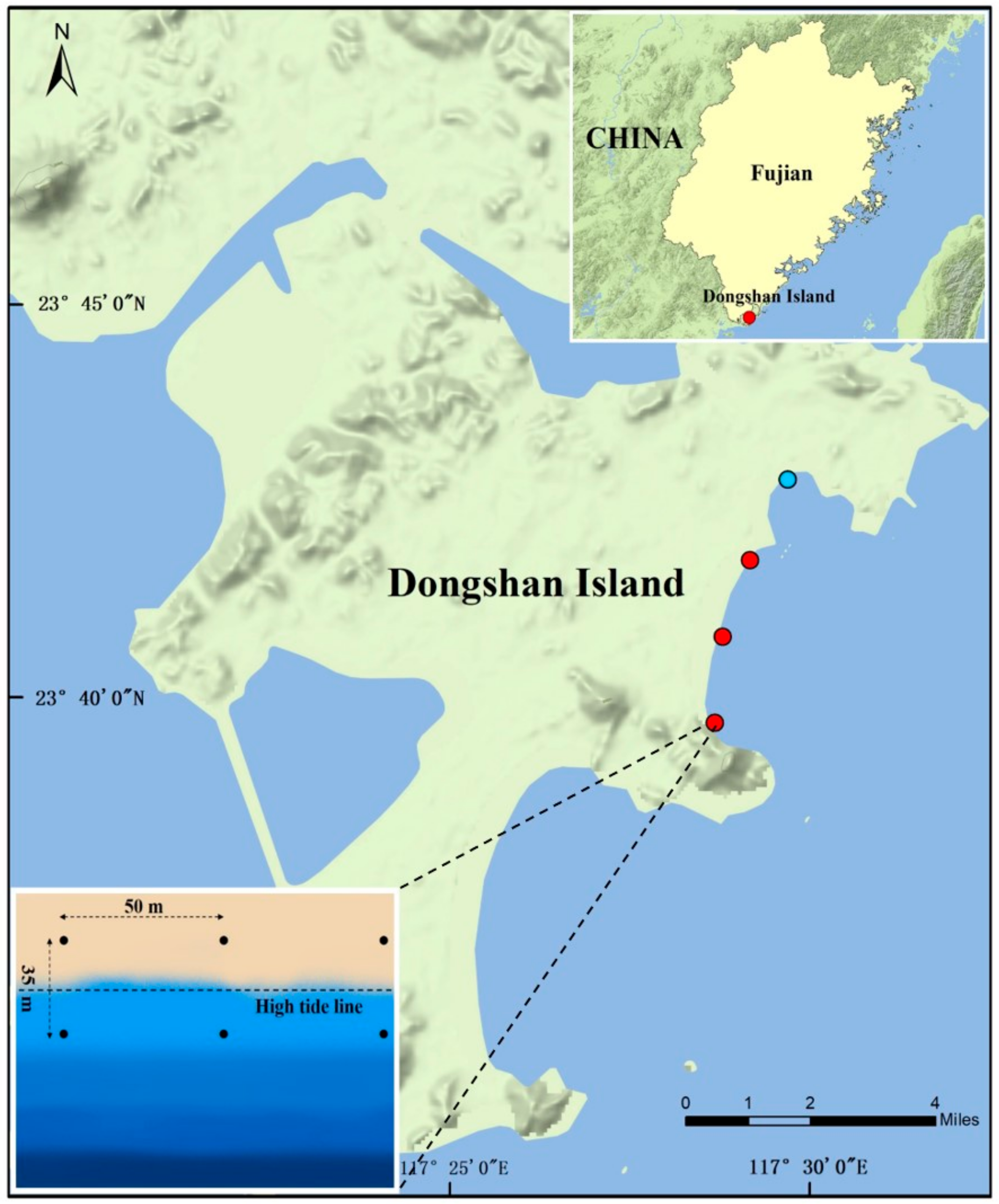
2.1. Sampling Area, Sampling Collection and Measurement of Environmental Parameters
2.2. DNA Extraction, PCR Amplification and 16S rDNA Amplicon Sequencing
2.3. Sequence Analysis
2.4. Statistical Analysis
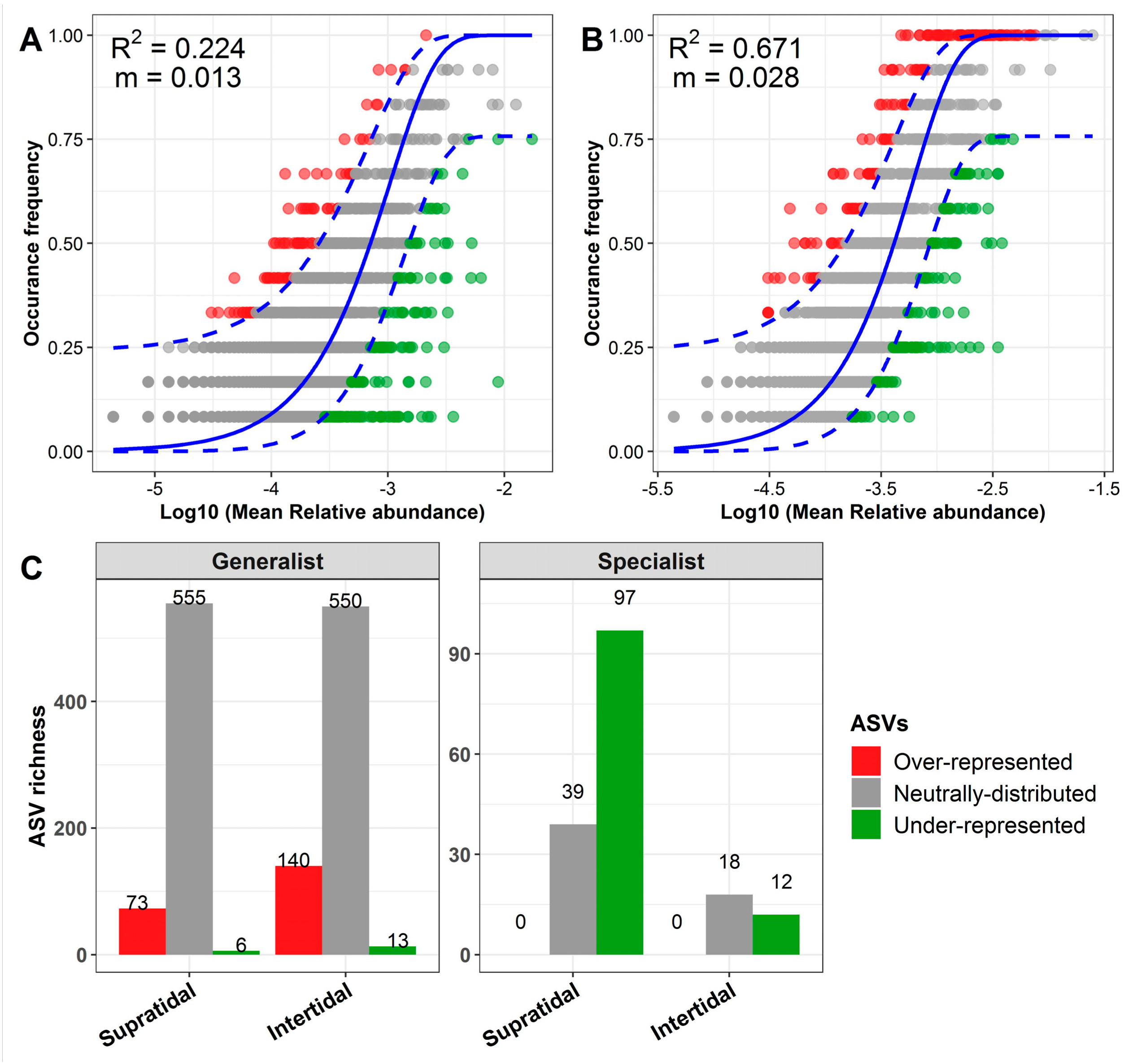
2.5. Null Model and Neutral Model Analysis
3. Results
3.1. Physico-Chemical Properties of the Coastal Sandy Sediments
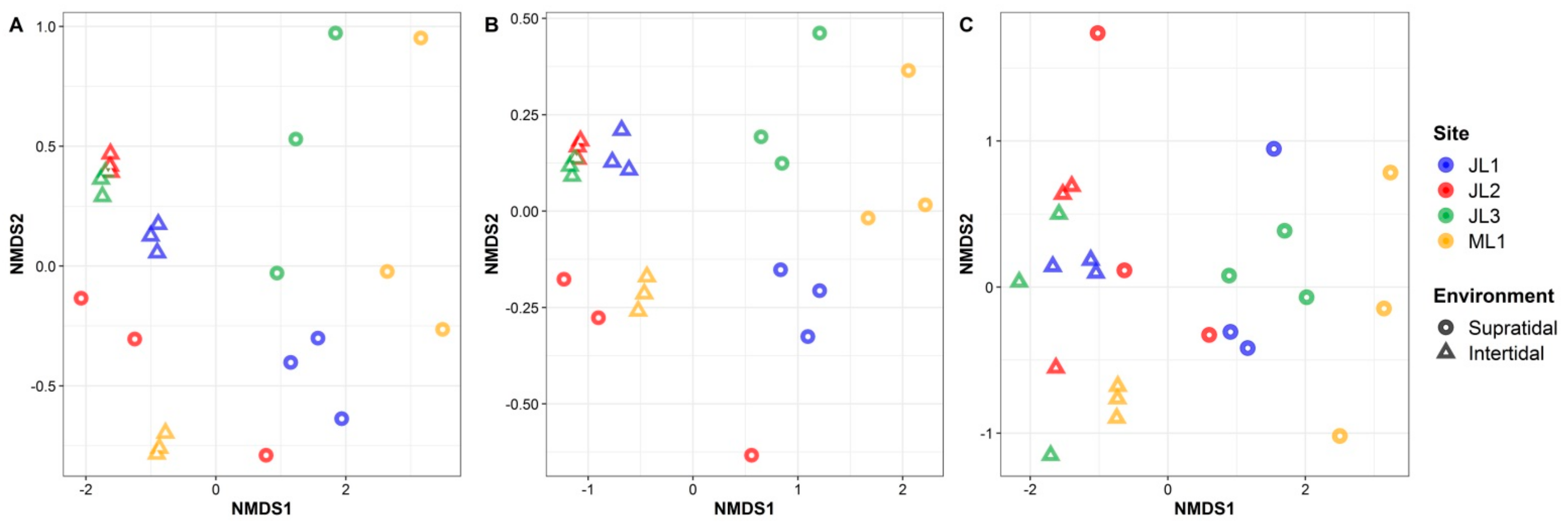
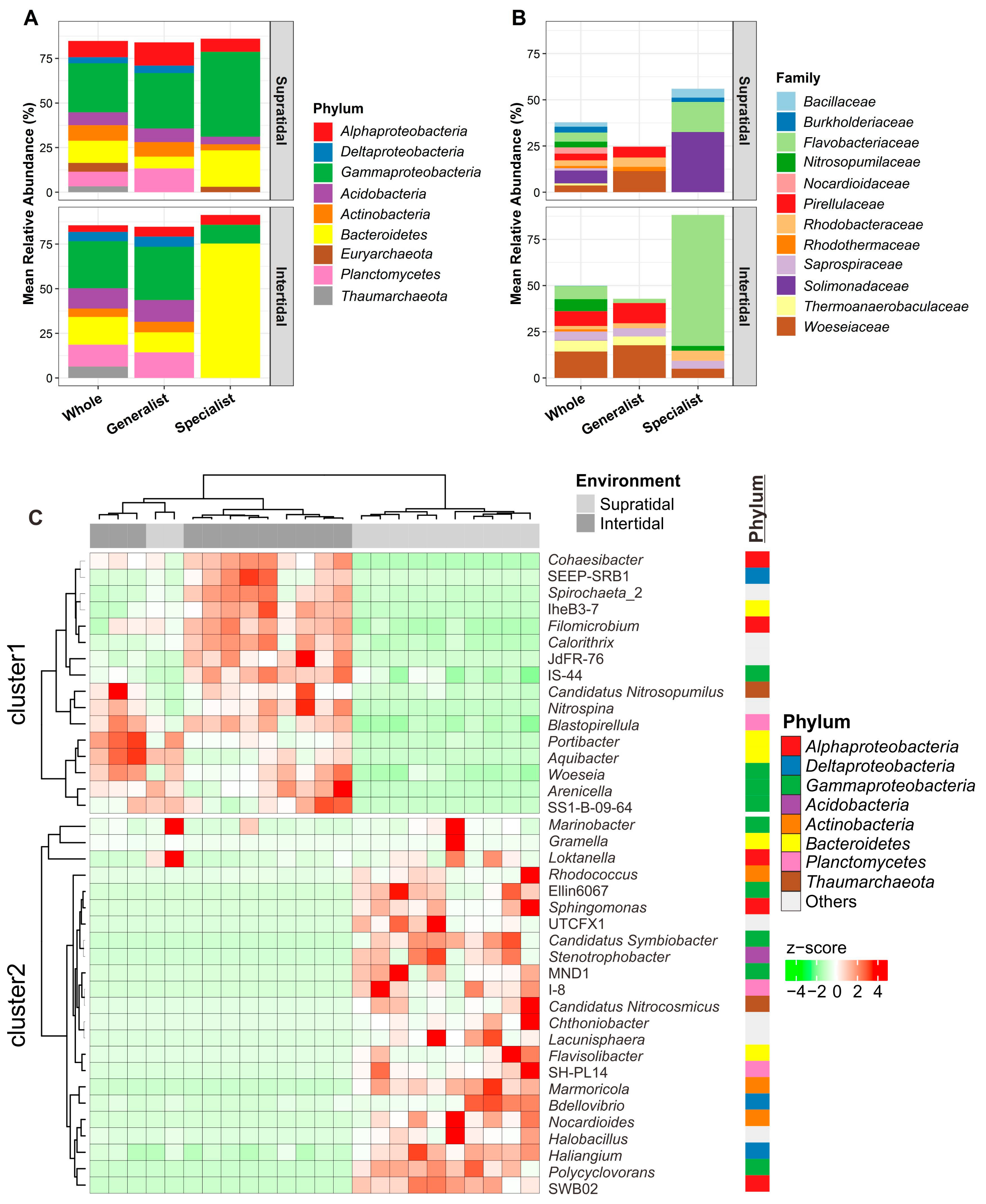
3.2. Variation in the Microbial Community Composition between the Supratidal and Intertidal Zones
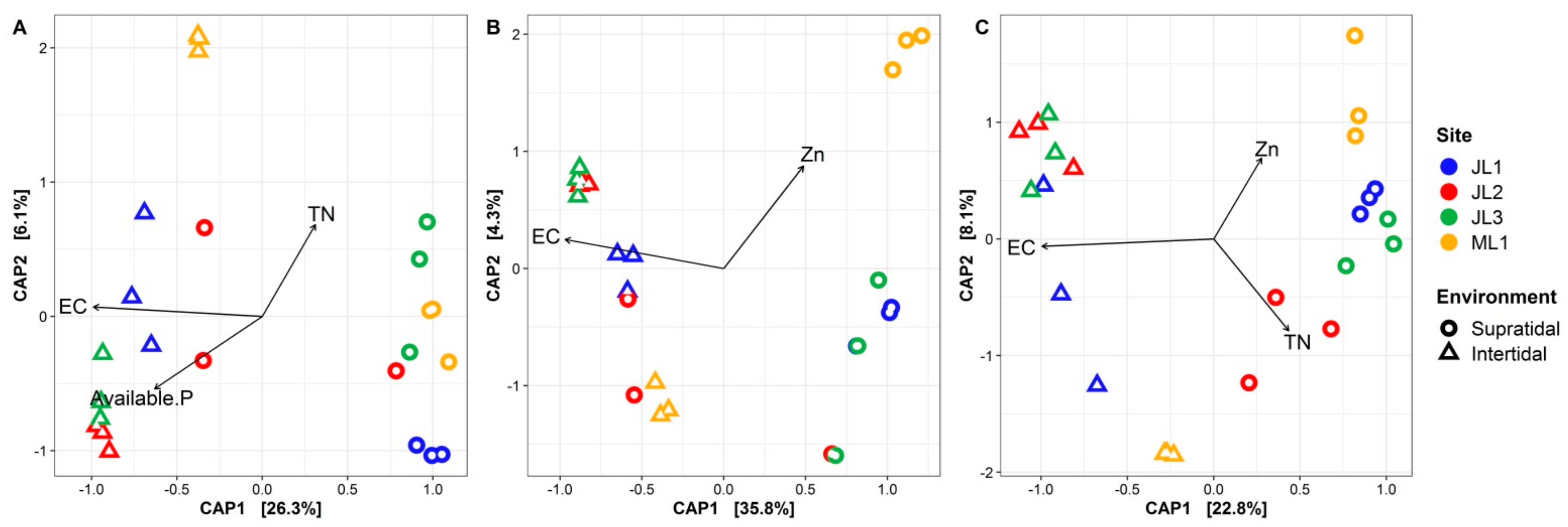
3.3. Relationship between Sand Microbial Communities and Environmental Factors
3.4. The Ecological Processes Underlying the Assembly of Sand Microbial Assemblages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lucrezi, S.; Schlacher, T.A.; Walker, S. Monitoring human impacts on sandy shore ecosystems: A test of ghost crabs (Ocypode spp.) as biological indicators on an urban beach. Environ. Monit. Assess. 2009, 152, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Yamahara, K.M.; Sassoubre, L.M. Diversity and transport of microorganisms in intertidal sands of the California coast. Appl. Environ. Microbiol. 2014, 80, 3943–3951. [Google Scholar] [CrossRef] [PubMed]
- Whitman, R.; Harwood, V.J.; Edge, T.A.; Nevers, M.; Byappanahalli, M.; Vijayavel, K.; Brandao, J.; Sadowsky, M.J.; Alm, E.W.; Crowe, A.; et al. Microbes in Beach Sands: Integrating Environment, Ecology and public health. Rev. Environ. Sci. Biotechnol. 2014, 13, 329–368. [Google Scholar] [CrossRef] [PubMed]
- Probandt, D.; Eickhorst, T.; Ellrott, A.; Amann, R.; Knittel, K. Microbial life on a sand grain: From bulk sediment to single grains. ISME J. 2018, 12, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Goto, D.; Yan, T. Effects of autochthonous microbial community on the die-off of fecal indicators in tropical beach sand. Fems. Microbiol. Ecol. 2010, 74, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Wanjugi, P.; Harwood, V.J. The influence of predation and competition on the survival of commensal and pathogenic fecal bacteria in aquatic habitats. Environ. Microbiol. 2013, 15, 517–526. [Google Scholar] [CrossRef]
- Staley, C.; Sadowsky, M.J. Regional Similarities and Consistent Patterns of Local Variation in Beach Sand Bacterial Communities throughout the Northern Hemisphere. Appl. Environ. Microbiol. 2016, 82, 2751–2762. [Google Scholar] [CrossRef]
- Zhang, W.; Pan, Y.; Yang, J.; Chen, H.; Holohan, B.; Vaudrey, J.; Lin, S.; McManus, G.B. The diversity and biogeography of abundant and rare intertidal marine microeukaryotes explained by environment and dispersal limitation. Environ. Microbiol. 2018, 20, 462–476. [Google Scholar] [CrossRef]
- Romao, D.; Staley, C.; Ferreira, F.; Rodrigues, R.; Sabino, R.; Verissimo, C.; Wang, P.; Sadowsky, M.; Brandao, J. Next-generation sequencing and culture-based techniques offer complementary insights into fungi and prokaryotes in beach sands. Mar. Pollu. Bull 2017, 119, 351–358. [Google Scholar] [CrossRef]
- Cloutier, D.D.; Alm, E.W.; McLellan, S.L. Influence of Land Use, Nutrients, and Geography on Microbial Communities and Fecal Indicator Abundance at Lake Michigan Beaches. Appl. Environ. Microbiol. 2015, 81, 4904–4913. [Google Scholar] [CrossRef]
- Boer, S.I.; Hedtkamp, S.I.; van Beusekom, J.E.; Fuhrman, J.A.; Boetius, A.; Ramette, A. Time- and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 2009, 3, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.L.; Overholt, W.A.; Hagan, C.; Huettel, M.; Kostka, J.E.; Konstantinidis, K.T. Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J. 2015, 9, 1928–1940. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, M.; Bian, X.; Guo, J.; Cai, L. A High-Level Fungal Diversity in the Intertidal Sediment of Chinese Seas Presents the Spatial Variation of Community Composition. Front. Microbiol. 2016, 7, 2098. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.I.; Boehm, A.B. Transport of Fecal Indicators from Beach Sand to the Surf Zone by Recirculating Seawater: Laboratory Experiments and Numerical Modeling. Environ. Sci. Technol. 2016, 50, 12840–12847. [Google Scholar] [CrossRef]
- Russell, T.L.; Yamahara, K.M.; Boehm, A.B. Mobilization and transport of naturally occurring enterococci in beach sands subject to transient infiltration of seawater. Environ. Sci. Technol. 2012, 46, 5988–5996. [Google Scholar] [CrossRef]
- Muller, E.E.L. Determining Microbial Niche Breadth in the Environment for Better Ecosystem Fate Predictions. mSystems 2019, 4, e00080–e00019. [Google Scholar] [CrossRef]
- Székely, A.J.; Langenheder, S. The importance of species sorting differs between habitat generalists and specialists in bacterial communities. FEMS Microbiol. Ecol. 2014, 87, 102–112. [Google Scholar] [CrossRef]
- Pandit, S.N.; Kolasa, J.; Cottenie, K. Contrasts between habitat generalists and specialists: An empirical extension to the basic metacommunity framework. Ecology 2009, 90, 2253–2262. [Google Scholar] [CrossRef]
- Peralta, A.L.; Matthews, J.W.; Kent, A.D. Habitat specialization along a wetland moisture gradient differs between ammonia-oxidizing and denitrifying microorganisms. Microb. Ecol. 2014, 68, 339–350. [Google Scholar] [CrossRef]
- Travis, J.M. Climate change and habitat destruction: A deadly anthropogenic cocktail. Proc. Biol. Sci. 2003, 270, 467–473. [Google Scholar] [CrossRef]
- Hou, L.; Hu, A.; Chen, S.; Zhang, K.; Orlic, S.; Rashid, A.; Yu, C.P. Deciphering the assembly processes of the key ecological assemblages of microbial communities in thirteen full-scale wastewater treatment plants. Microbes. Environ. 2019, 34, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Cao, X.; Zhao, L.; Wang, J.; Gao, Z.; Wang, M.C.; Huang, Y. The importance of neutral and niche processes for bacterial community assembly differs between habitat generalists and specialists. FEMS Microbiol. Ecol. 2016, 92, fiw174. [Google Scholar] [CrossRef] [PubMed]
- Lindh, M.V.; Sjostedt, J.; Casini, M.; Andersson, A.; Legrand, C.; Pinhassi, J. Local environmental conditions shape generalist but not specialist components of microbial metacommunities in the baltic sea. Front. Microbiol. 2016, 7, 2078. [Google Scholar] [CrossRef] [PubMed]
- Weiskerger, C.J.; Brandao, J.; Ahmed, W.; Aslan, A.; Avolio, L.; Badgley, B.D.; Boehm, A.B.; Edge, T.A.; Fleisher, J.M.; Heaney, C.D.; et al. Impacts of a changing earth on microbial dynamics and human health risks in the continuum between beach water and sand. Water Res. 2019, 162, 456–470. [Google Scholar] [CrossRef]
- Tucker, C.M.; Shoemaker, L.G.; Davies, K.F.; Nemergut, D.R.; Melbourne, B.A. Differentiating between niche and neutral assembly in metacommunities using null models of β-diversity. Oikos 2016, 125, 778–789. [Google Scholar] [CrossRef]
- Sloan, W.T.; Lunn, M.; Woodcock, S.; Head, I.M.; Nee, S.; Curtis, T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 2006, 8, 732–740. [Google Scholar] [CrossRef]
- Hu, A.; Ju, F.; Hou, L.; Li, J.; Yang, X.; Wang, H.; Mulla, S.I.; Sun, Q.; Bürgmann, H.; Yu, C.-P. Strong impact of anthropogenic contamination on the co-occurrence patterns of a riverine microbial community. Environ. Microbiol. 2017, 19, 4993–5009. [Google Scholar] [CrossRef]
- Hu, A.; Yang, X.; Chen, N.; Hou, L.; Ma, Y.; Yu, C.-P. Response of bacterial communities to environmental changes in a mesoscale subtropical watershed, Southeast China. Sci. Total Environ. 2014, 472, 746–756. [Google Scholar] [CrossRef]
- Olsen, S.R.; Cole, C.V. Estimation of available P in soils by extraction with sodium bicarbonate. USDA Circ. 1954, 939, 18–19. [Google Scholar]
- Quince, C.; Lanzen, A.; Davenport, R.J.; Turnbaugh, P.J. Removing noise from pyrosequenced amplicons. BMC Bioinform. 2011, 12, 38. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Feld, C.K.; Segurado, P.; Gutierrez-Canovas, C. Analysing the impact of multiple stressors in aquatic biomonitoring data: A ‘cookbook’ with applications in R. Sci. Total Environ. 2016, 573, 1320–1339. [Google Scholar] [CrossRef] [PubMed]
- Legendre, P.; Mi, X.C.; Ren, H.B.; Ma, K.P.; Yu, M.J.; Sun, I.F.; He, F.L. Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 2009, 90, 663–674. [Google Scholar] [CrossRef] [Green Version]
- Borcard, D.; Legendre, P. All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecol. Model. 2002, 153, 51–68. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Simpson, G.; Solymos, P.; Stevens, M.; Wagner, H. Vegan: Community Ecology Package. R package version 1.15-2. Available online: http://cran.r-project.org/web/packages/vegan/index.html.
- Roberts, D. Labdsv: Ordination and multivariate analysis for ecology. R Package Version 2007, 1. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant graphics for data analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Gu, Z.G.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Burns, A.R.; Stephens, W.Z.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016, 10, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.; Dai, W.; Qiu, Q.; Zhu, J.; Yang, W.; Li, C. Response of host-bacterial colonization in shrimp to developmental stage, environment and disease. Mol. Ecol. 2018, 27, 3686–3699. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, A.; Hou, L.; Yu, C.P. Biogeography of planktonic and benthic archaeal communities in a subtropical eutrophic estuary of China. Microb. Ecol. 2015, 70, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Herlemann, D.P.R.; Labrenz, M.; Jurgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Auguet, J.C.; Barberan, A.; Casamayor, E.O. Global ecological patterns in uncultured Archaea. ISME J. 2010, 4, 182–190. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Shi, Y.; Cui, X.; Yue, P.; Li, K.; Liu, X.; Tripathi, B.M.; Chu, H. Salinity Is a Key Determinant for Soil Microbial Communities in a Desert Ecosystem. mSystems 2019, 4, e00225–e00218. [Google Scholar] [CrossRef] [Green Version]
- Dupont, C.L.; Larsson, J.; Yooseph, S.; Ininbergs, K.; Goll, J.; Asplund-Samuelsson, J.; McCrow, J.P.; Celepli, N.; Allen, L.Z.; Ekman, M.; et al. Functional Tradeoffs Underpin Salinity-Driven Divergence in Microbial Community Composition. PLoS ONE 2014, 9, e89549. [Google Scholar] [CrossRef]
- Henson, M.W.; Lanclos, V.C.; Faircloth, B.C.; Thrash, J.C. Cultivation and genomics of the first freshwater SAR11 (LD12) isolate. ISME J. 2018, 12, 1846–1860. [Google Scholar] [CrossRef] [Green Version]
- Bowman, J.P. The Marine Clade of the Family Flavobacteriaceae: The Genera Aequorivita, Arenibacter, Cellulophaga, Croceibacter, Formosa, Gelidibacter, Gillisia, Maribacter, Mesonia, Muricauda, Polaribacter, Psychroflexus, Psychroserpens, Robiginitalea, Salegentibacter, Tenacibaculum, Ulvibacter, Vitellibacter and Zobellia. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 677–694. [Google Scholar]
- Zhou, Y.; Lai, R.; Li, W.-J. The Family Solimonadaceae. In The Prokaryotes; Rosenberg, E., De Long, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: New York, NY, USA, 2014; pp. 627–638. [Google Scholar]
- Walker, C.B.; de la Torre, J.R.; Klotz, M.G.; Urakawa, H.; Pinel, N.; Arp, D.J.; Brochier-Armanet, C.; Chain, P.S.G.; Chan, P.P.; Gollabgir, A. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. USA. 2010, 107, 8818–8823. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.Y.; Kim, J.G.; Sinninghe Damste, J.S.; Rijpstra, W.I.; Madsen, E.L.; Kim, S.J.; Hong, H.; Si, O.J.; Kerou, M.; Schleper, C.; et al. A hydrophobic ammonia-oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar-contaminated sediment. Environ. Microbiol. Rep. 2016, 8, 983–992. [Google Scholar] [CrossRef]
- Gobet, A.; Boer, S.I.; Huse, S.M.; van Beusekom, J.E.; Quince, C.; Sogin, M.L.; Boetius, A.; Ramette, A. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 2012, 6, 542–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lear, G.; Bellamy, J.; Case, B.S.; Lee, J.E.; Buckley, H.L. Fine-scale spatial patterns in bacterial community composition and function within freshwater ponds. ISME J. 2014, 8, 1715–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; Mulla, S.I.; Nino-Garcia, J.P.; Ning, D.; Rashid, A.; Hu, A.; Yu, C.P. Deterministic and stochastic processes driving the shift in the prokaryotic community composition in wastewater treatment plants of a coastal Chinese city. Appl. Microbiol. Biotechnol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ren, K.; Isabwe, A.; Chen, H.; Liu, M.; Yang, J. Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome 2019, 7, 138. [Google Scholar] [CrossRef] [Green Version]
- Chase, J.M.; Kraft, N.J.B.; Smith, K.G.; Vellend, M.; Inouye, B.D. Using null models to disentangle variation in community dissimilarity from variation in alpha-diversity. Ecosphere 2011, 2, 1–11. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, W.; Deng, Y.; Jiang, Y.H.; Xue, K.; He, Z.; Van Nostrand, J.D.; Wu, L.; Yang, Y.; Wang, A. Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. MBio 2013, 4, e00584–e00512. [Google Scholar] [CrossRef] [Green Version]
- Yao, Z.; Du, S.; Liang, C.; Zhao, Y.; Dini-Andreote, F.; Wang, K.; Zhang, D. Bacterial community assembly in a typical estuarine marsh with multiple environmental gradients. Appl. Environ. Microbiol. 2019, 85, e02602–e02618. [Google Scholar] [CrossRef] [Green Version]
- Finlay, B.J.; Fenchel, T. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist 2004, 155, 237–244. [Google Scholar] [CrossRef]


| Group | Factor a | Adonis | ANOSIM | ||
|---|---|---|---|---|---|
| R2 | p | R | p | ||
| Whole | Environment | 0.263 | <0.001 | 0.669 | <0.001 |
| Site | 0.200 | <0.001 | 0.131 | 0.063 | |
| E × S | 0.195 | <0.001 | — | — | |
| Habitat generalists | Environment | 0.337 | <0.001 | 0.641 | <0.001 |
| Site | 0.205 | <0.001 | 0.137 | 0.061 | |
| E × S | 0.194 | <0.001 | — | — | |
| Habitat specialists | Environment | 0.236 | <0.001 | 0.686 | <0.001 |
| Site | 0.178 | <0.001 | 0.062 | 0.178 | |
| E × S | 0.203 | <0.001 | — | — | |
| Groups | Whole | Habitat Generalists | Habitat Specialists | |
|---|---|---|---|---|
| Partial db-RDA | Pure Env a | 10.5% **b | 11.8% *** | 7.4% ** |
| Pure Spat | 6.7% ** | 9.7% ** | 8.8% ** | |
| Shared | 17.3% | 19.1% | 18.8% | |
| Total | 34.5% *** | 40.5% *** | 35.0% *** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, A.; Wang, H.; Cao, M.; Rashid, A.; Li, M.; Yu, C.-P. Environmental Filtering Drives the Assembly of Habitat Generalists and Specialists in the Coastal Sand Microbial Communities of Southern China. Microorganisms 2019, 7, 598. https://doi.org/10.3390/microorganisms7120598
Hu A, Wang H, Cao M, Rashid A, Li M, Yu C-P. Environmental Filtering Drives the Assembly of Habitat Generalists and Specialists in the Coastal Sand Microbial Communities of Southern China. Microorganisms. 2019; 7(12):598. https://doi.org/10.3390/microorganisms7120598
Chicago/Turabian StyleHu, Anyi, Hongjie Wang, Meixian Cao, Azhar Rashid, Mingfeng Li, and Chang-Ping Yu. 2019. "Environmental Filtering Drives the Assembly of Habitat Generalists and Specialists in the Coastal Sand Microbial Communities of Southern China" Microorganisms 7, no. 12: 598. https://doi.org/10.3390/microorganisms7120598





