Hyperspectral Imaging to Evaluate the Effect of IrrigationWater Salinity in Lettuce
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Analytical Measurements
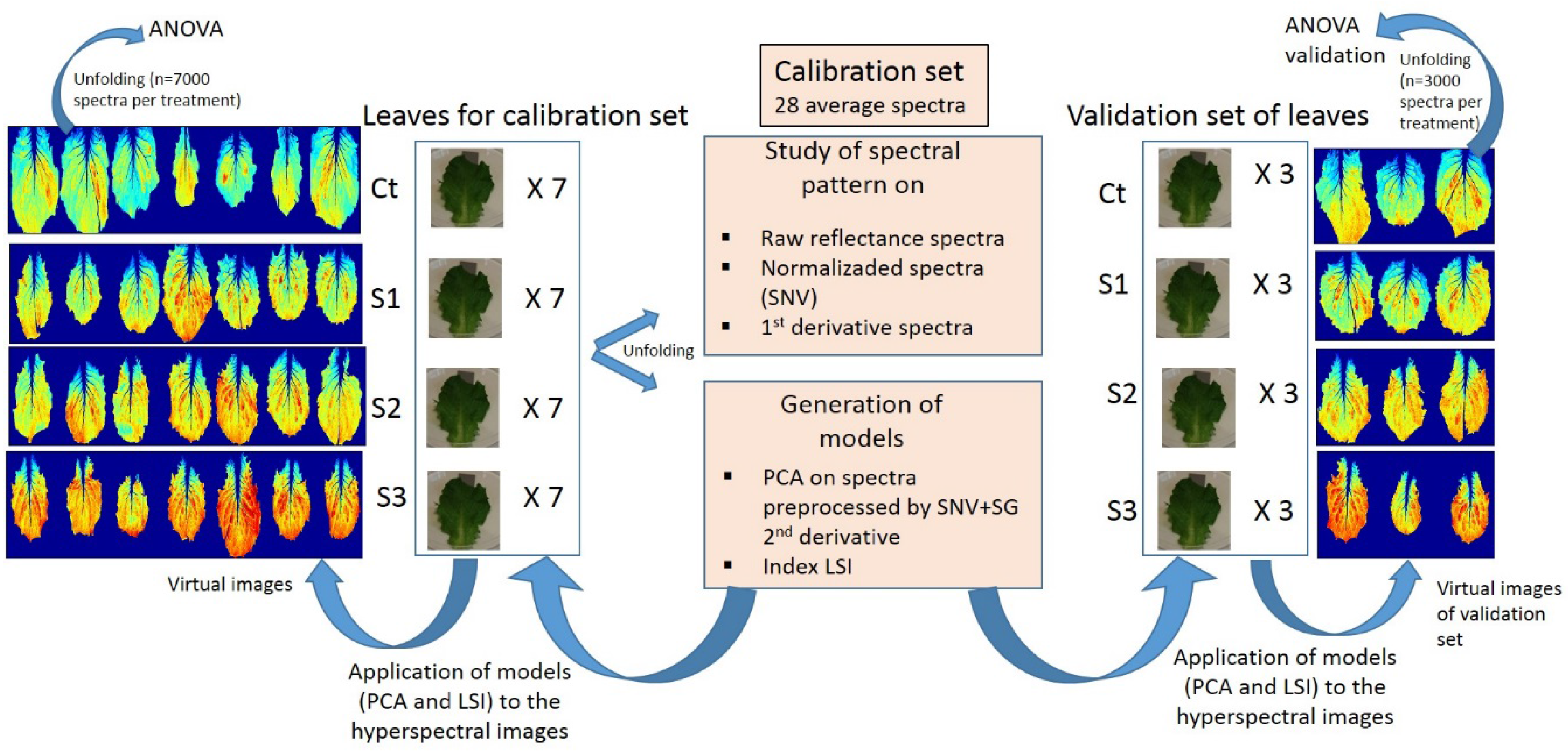
2.2. Hyperspectral Images
Data Preprocessing and Processing. Computation of Models
3. Results and Discussion
3.1. Effect of Salinity on Water Content and Osmotic Potential
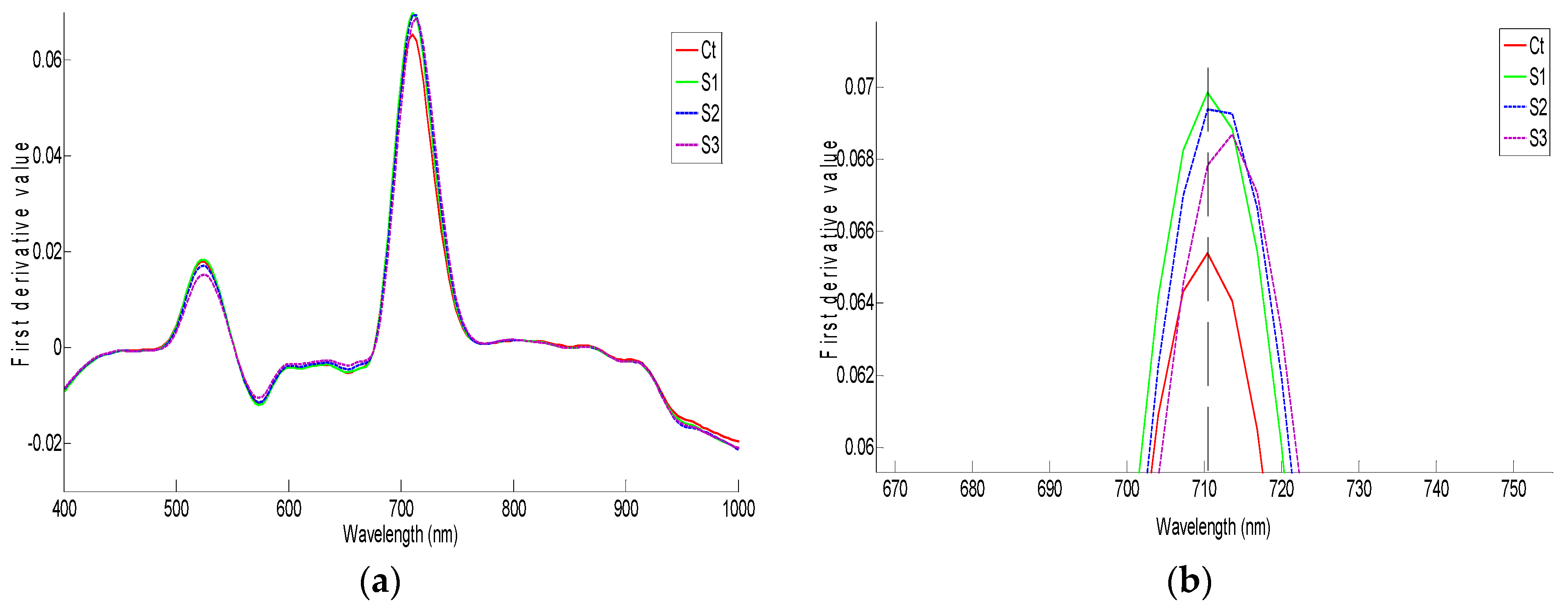
3.2. Effect of Salinity on Spectral Features
3.2.1. Non Preprocessed and SNV Reflectance Spectra
3.2.2. First Derivative of the Spectra
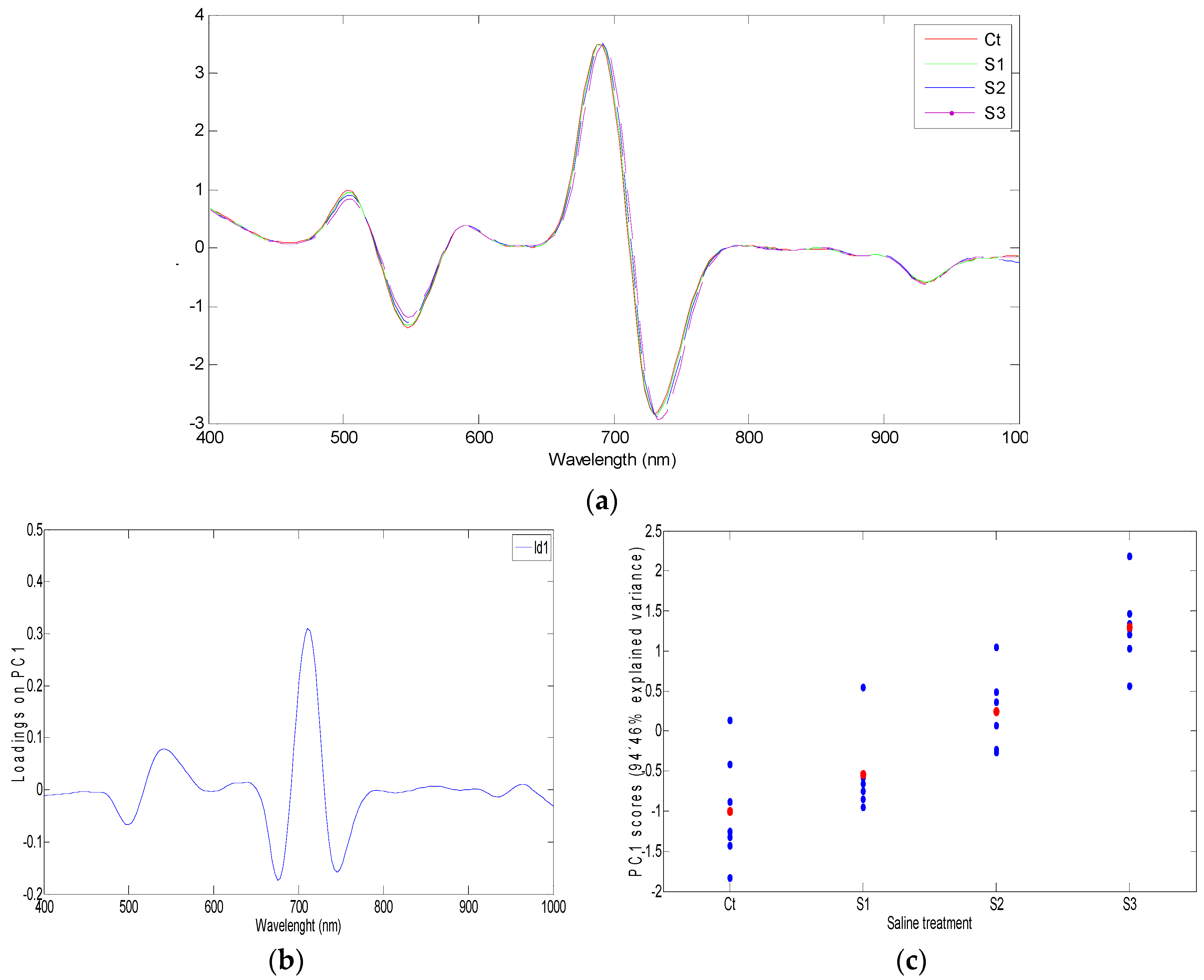
3.3. Models and Data Processing
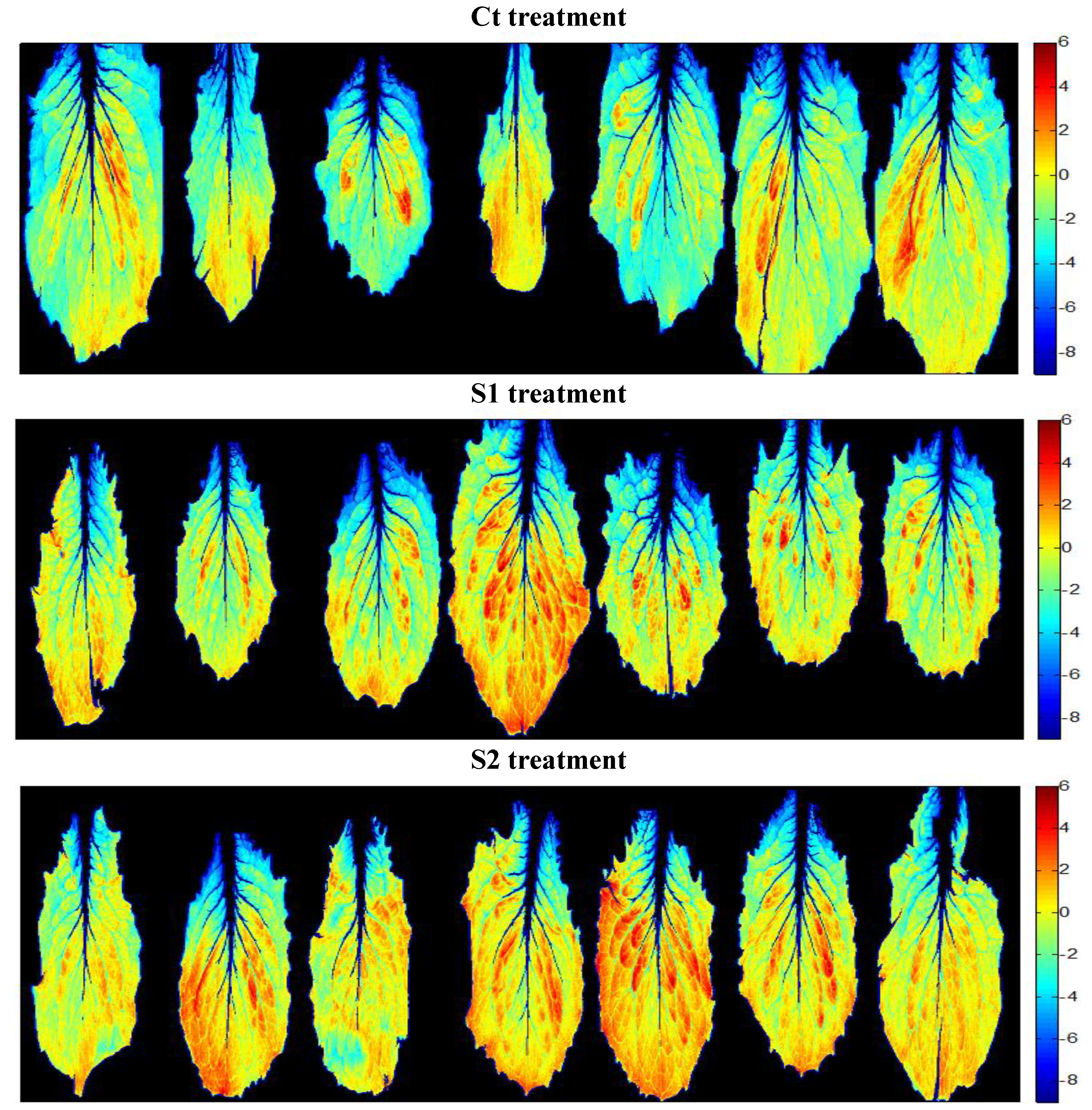
3.3.1. Second Derivative and SNV Normalization: Principal Component Analysis
3.3.2. A New Index on the Red Edge Region: Level Salinity Index LSI
3.3.3. Comparison between Spectral Indexes
3.4. Validation of the Models to Evaluate Salinity Effects
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lamsal, K.; Paudyal, G.N.; Saeed, M. Model for assessing impact of salinity on soil water availability and crop yield. Agric. Water Manag. 1999, 41, 57–70. [Google Scholar] [CrossRef]
- Shannon, M.C.; Grieve, C.M.; Francois, L.E. Whole-plant response to salinity. In Plant-Environment Interactions; Wilkinson, R.E., Ed.; Marcel Dekker: New York, NY, USA, 1994; pp. 199–244. [Google Scholar]
- Acosta, J.A.; Faz, A.; Jansen, B.; Kalbitz, K.; Martínez-Martínez, S. Assessment of salinity status in intensively cultivated soils under semiarid climate, Murcia, SE Spain. J. Arid Environ. 2011, 75, 1056–1066. [Google Scholar] [CrossRef]
- Ünlükara, A.; Cemek, B.; Karaman, S.; Erşahin, S. Response of lettuce (Lactuca sativa var. crispa) to salinity of irrigation water. N. Z. J. Crop Horticult. Sci. 2008, 36, 265–273. [Google Scholar]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Maas, E.V.; Hoffman, G.J. Crop salt tolerance ± current assessment. J. Irrig. Drain. Eng. 1977, 103, 115–134. [Google Scholar]
- Bie, Z.; Ito, T.; Shinohara, Y. Effects of sodium sulfate and sodium bicarbonate on the growth, gas exchange and mineral composition of lettuce. Sci. Horticult. 2004, 99, 215–224. [Google Scholar] [CrossRef]
- Eraslan, F.; Inal, A.; Savasturk, O.; Gunes, A. Changes in antioxidative system and membrane damage of lettuce in response to salinity and boron toxicity. Sci. Horticult. 2007, 114, 5–10. [Google Scholar] [CrossRef]
- Carassay, L.R.; Bustos, D.A.; Golberg, A.D.; Taleisnik, E. Tipburn in salt-affected lettuce (Lactuca sativa L.) plants results from local oxidative stress. J. Plant Physiol. 2012, 169, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.; Adams, P. Diagnosis of mineral disorders in plants. In Glasshouse Crops; Robinson, J.B.D., Ed.; Ministry of Agriculture, Fisheries and Food/Agriculture and Fisheries Research Council, HMSO: London, UK, 1987. [Google Scholar]
- Poss, J.A.; Grattan, S.R.; Grieve, C.M.; Shannon, M.C. Characterization of leaf boron injury in salt-stressed Eucalyptus by image analysis. Plant Soil 1999, 206, 237–245. [Google Scholar] [CrossRef]
- Saure, M.C. Causes of the tipburn disorder in leaves of vegetables. Sci. Horticult. 1998, 76, 131–147. [Google Scholar] [CrossRef]
- Poss, J.A.; Russell, W.B.; Grieve, C.M. Estimating yields of salt- and water-stressed forages with remote sensing in the visible and near infrared. J. Environ. Qual. 2006, 35, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Hamzeh, S.; Naseri, A.A.; AlaviPanah, S.K.; Mojaradi, B.; Bartholomeus, H.M.; Clevers, J.G.P.W.; Behzad, M. Estimating salinity stress in sugarcane fields with spaceborne hyperspectral vegetation indices. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 282–290. [Google Scholar] [CrossRef]
- Zhang, T.; Zeng, S.; Gao, Y.; Ouyang, Z.; Li, B.; Fang, C.; Zhao, B. Using hyperspectral vegetation indices as a proxy to monitor soil salinity. Ecol. Indic. 2011, 11, 1552–1562. [Google Scholar] [CrossRef]
- Tucker, C.J. Remote sensing of leaf water content in the near infrared. Remote Sens. Environ. 1980, 10, 23–32. [Google Scholar] [CrossRef]
- Harris, A.; Bryant, R.G.; Baird, A.J. Mapping the effects of water stress on Sphagnum: Preliminary observations using airborne remote sensing. Remote Sens. Environ. 2006, 100, 363–378. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Kooistra, L.; Schaepman, M.E. Estimating canopy water content using hyperspectral remote sensing data. Int. J. Appl. Earth Obs. Geoinf. 2010, 12, 119–125. [Google Scholar] [CrossRef]
- Suárez, L.; Zarco-Tejada, P.J.; González-Dugo, V.; Berni, J.A.J.; Sagardoy, R.; Morales, F.; Fereres, E. Detecting water stress effects on fruit quality in orchards with time-series PRI airborne imagery. Remote Sens. Environ. 2010, 114, 286–298. [Google Scholar] [CrossRef]
- Al-Abbas, A.H.; Barr, R.; Hall, J.D.; Crance, F.L.; Baumgardner, M.F. Spectra of normal and nutrient-deficient maize leaves. Agron. J. 1974, 66, 16–20. [Google Scholar] [CrossRef]
- Masoni, A.; Ercoli, L.; Mariotti, M. Spectral properties of leaves deficient in iron, sulfur, magnesium, and manganese. Agron. J. 1996, 88, 937–943. [Google Scholar] [CrossRef]
- Pacumbaba, R.O., Jr.; Beyl, C.A. Changes in hyperspectral reflectance signatures of lettuce leaves in response to macronutrient deficiencies. Adv. Space Res. 2011, 48, 32–42. [Google Scholar] [CrossRef]
- Agüero, M.V.; Ponce, A.G.; Moreira, M.R.; Roura, S.I. Lettuce quality loss under conditions that favor the wilting phenomenon. Postharvest Biol. Technol. 2011, 59, 124–131. [Google Scholar] [CrossRef]
- Van’t Hoff, J.H. The role of osmotic pressure in the analogy between solution and gases. Z. Phys. Chem. 1887, 1, 481–508. [Google Scholar] [CrossRef]
- Richardson, A.D.; Duigan, S.P.; Berlyn, G.P. An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol. 2002, 153, 185–194. [Google Scholar] [CrossRef]
- Li, G.; Wan, S.; Zhou, J.; Yang, Z.; Qin, P. Leaf chlorophyll fluorescence, hyperspectral reflectance, pigments content, malondialdehyde and proline accumulation responses of castor bean (Ricinus communis L.) seedlings to salt stress levels. Ind. Crops Prod. 2010, 31, 13–19. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Lara, M.A.; Lleó, L.; Diezma-Iglesias, B.; Roger, J.M.; Ruiz-Altisent, M. Monitoring spinach shelf-life with hyperspectral image through packaging films. J. Food Eng. 2013, 119, 353–361. [Google Scholar] [CrossRef] [Green Version]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Blum, A. Plant Breeding for Stress Environment; CRC Press Inc.: Boca Raton, FL, USA, 1988. [Google Scholar]
- Leidi, E.O.; Pardo, J.M. Tolerancia de los cultivos al estrés salino: Qué hay de nuevo. Revista de Investigaciones de la Facultad de Ciencias Agrarias 2002, UNR 2, 70–91. [Google Scholar]
- Gitelson, A.; Viña, A.; Arkebauer, T.J.; Rundquist, D.C.; Keydan, G.; Leavitt, B. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30, 1248. [Google Scholar] [CrossRef]
- Knipling, E.B. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.; Zur, Y.; Chivkunova, O.B.; Merzlyak, M.N. Assessing carotenoid content in plant leaves with reflectance spectroscopy. Photochem. Photobiol. 2002, 75, 272–281. [Google Scholar] [CrossRef]
- Peñuelas, P.; Isla, R.; Filella, I.; Araus, J.L. Visible and near infrared reflectance assessment of salinity effects on barley. Crop Sci. 1997, 37, 198–202. [Google Scholar] [CrossRef]
- Wang, D.; Wilson, C.; Shannon, M.C. Interpretation of salinity and irrigation effects on soybean canopy reflectance in visible and near-infrared spectrum domain. Int. J. Remote Sens. 2002, 23, 811–824. [Google Scholar] [CrossRef]
- Mariotti, M.; Ercoli, L.; Masoni, A. Spectral properties of iron deficient corn and sunflower leaves. Remote Sens. Environ. 1996, 58, 282–288. [Google Scholar] [CrossRef]
- Ustin, S.L.; Gitelson, A.A.; Jacquemoud, S.; Schaepman, M.; Asner, G.P.; Gamon, J.A.; Zarco-Tejada, P. Retrieval of foliar information about plant pigment systems from high resolution spectroscopy. Remote Sens. Environ. 2009, 113, S67–S77. [Google Scholar] [CrossRef] [Green Version]
- Peñuelas, J.; Pinol, J.; Ogaya, R.; Filella, I. Estimation of plant water concentration by the reflectance water index wi (r900/r970). Int. J. Remote Sens. 1997, 18, 2869–2875. [Google Scholar] [CrossRef]
- Diezma, B.; Lleó, L.; Roger, J.M.; Herrero-Langreo, A.; Lunadei, L.; Ruiz-Altisent, M. Examination of the quality of spinach leaves using hyperspectral imaging. Postharvest Biol. Technol. 2013, 85, 8–17. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Spectral reflectance changes associated with autumn senescence of Aesculus hippocastanum L. and Acer platanoides. Leaves spectal features and relation to chlorophyll estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N.; Lichtenthaler, H.K. Detection of red edge position and chlorophyll content by reflectance measurements near 700 nm. J. Plant Physiol. 1996, 148, 501–508. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Keydan, G.P.; Merzlyak, M.N. Three-Band Model for Non-Invasive Estimation of Chlorophyll Carotenoids and Anthocyanin Contents in Higher Plant Leaves. Papers in Natural Resources. 2006. Paper 258. Available online: http://digitalcommons.unl.edu/natrespapers/258 (accessed on 1 November 2016).
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Rud, R.; Shoshany, M.; Alchanatis, V. Spectral indicators for salinity effects in crops: A comparison of a new green indigo ratio with existing indices. Remote Sens. Lett. 2011, 2, 289–298. [Google Scholar] [CrossRef]
- Holbrook, N.M. El balance hídrico de las plantas. In Fisología Vegetal; Taiz, L., Zeiger, E., Eds.; Universitat Jaume I: Castelló de la Plana, Spain, 2006; pp. 79–115. [Google Scholar]
- Lazof, D.B.; Bernstein, N. Effects of salinization on nutrient transport to lettuce leaves: Consideration of leaf developmental stage. New Phytol. 1999, 144, 85–94. [Google Scholar] [CrossRef]


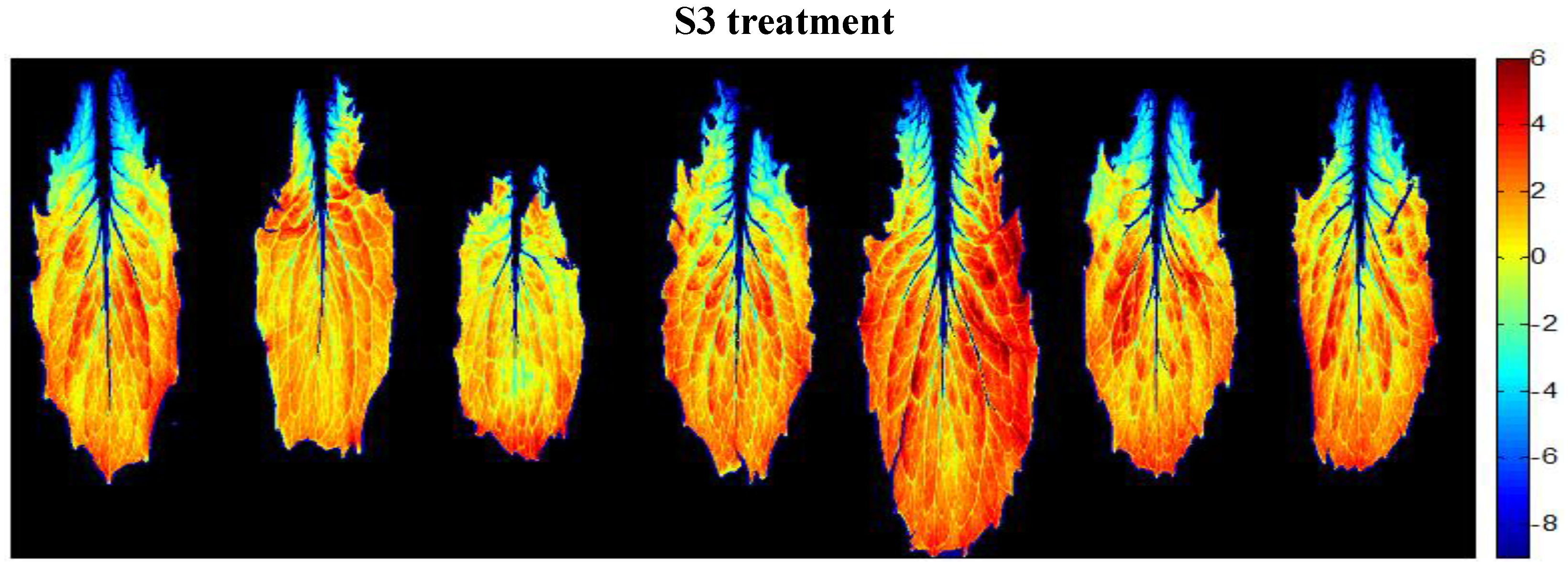
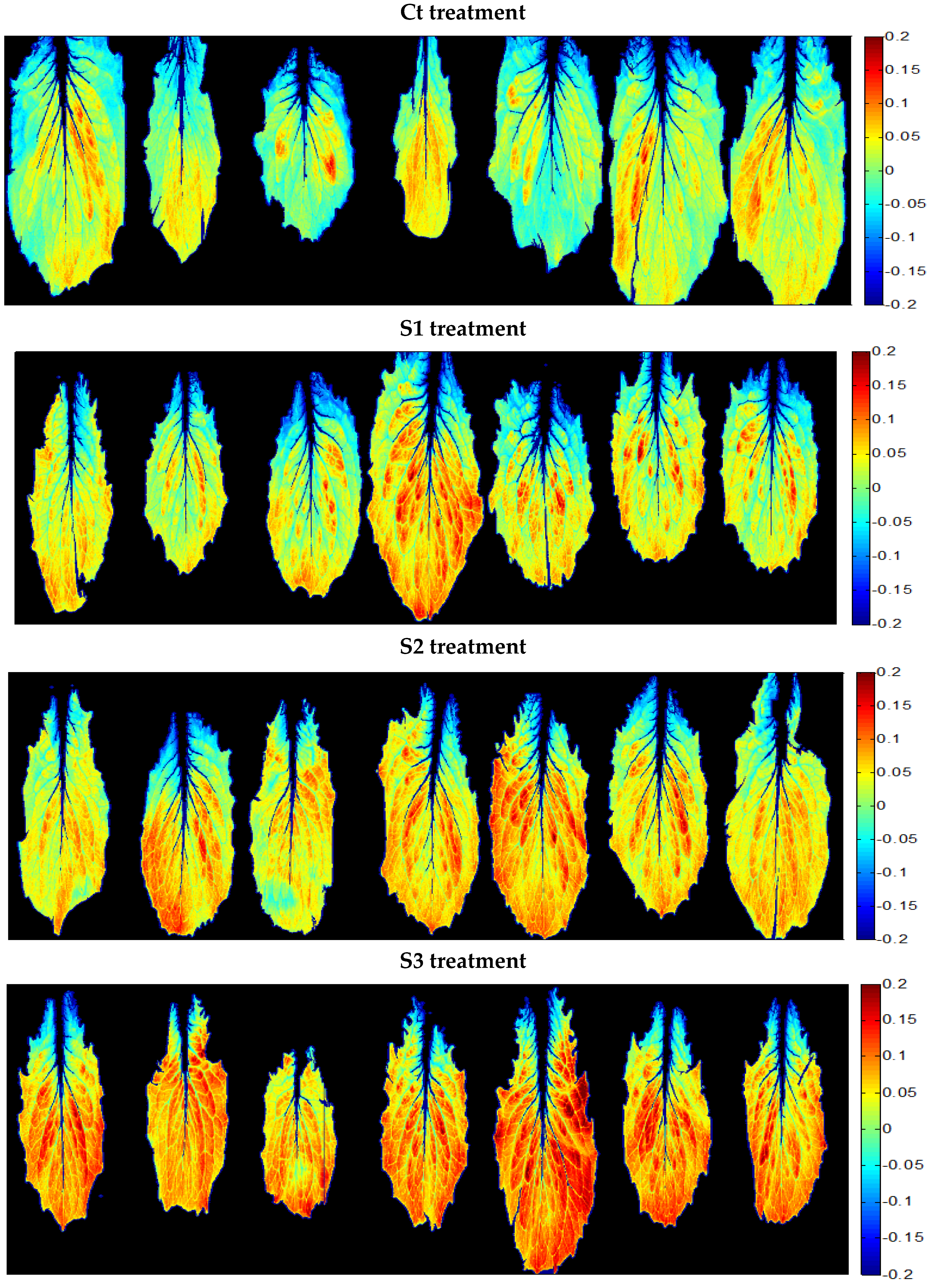

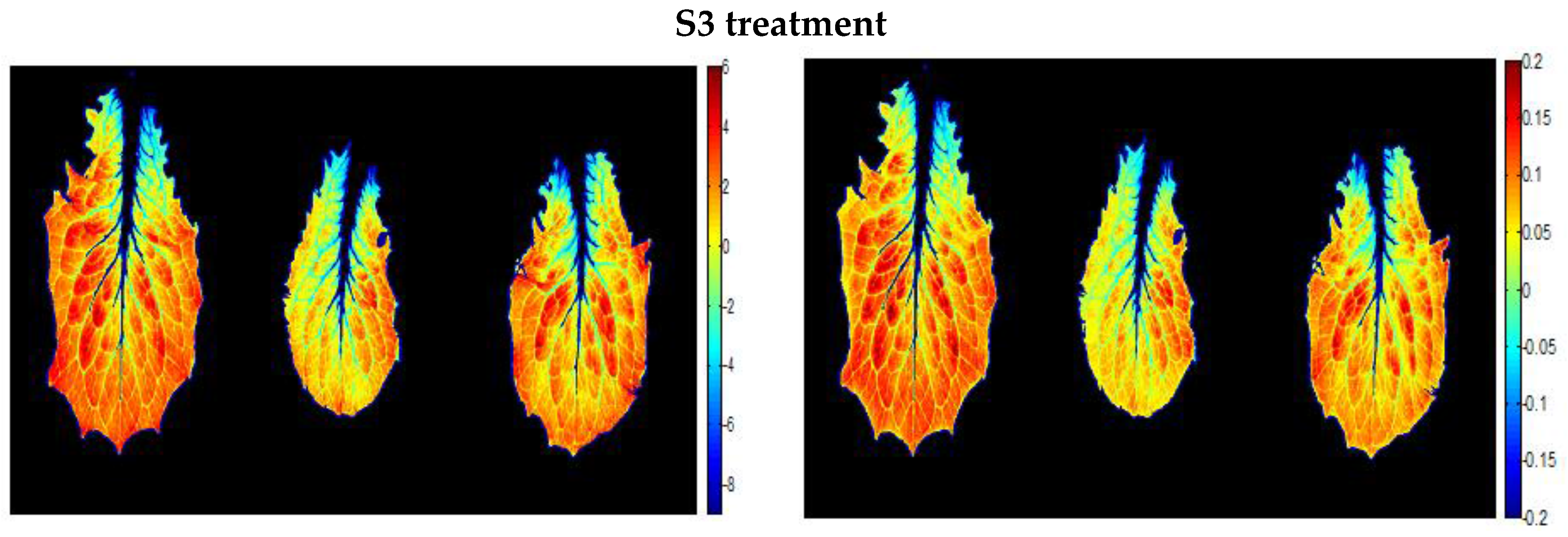
| Saline Treatment | Water Content (%) | Osmotic Potential (MPa) |
|---|---|---|
| Mean ± STD | Mean ± STD | |
| Ct | 94.53 ± 0.09a | −0.65 ± 0.04a |
| S1 | 94.54 ± 0.12a | −1.08 ± 0.09b |
| S2 | 93.15 ± 0.18b | −1.49 ± 0.11c |
| S3 | 92.70 ± 0.07c | −2.06 ± 0.09d |
| Index | Formula |
|---|---|
| NDVI, related to chlorophyll content [42] | NDVI = (R800 − R670)/(R800 + R670) |
| NDVI705, related to chlorophyll content [43] | NDVI705 = (R750 − R705)/(R750 + R705) |
| Red Edge Position Index, related to chlorophyll content [44] | RPI = (R750/R700) |
| Related to chlorophyll content in anthocyanin free leaves [45] | Chlgreen = (R760/R550) − 1 Chlred edge = (R760/R705) − 1 |
| Plant Senescence Reflectance Index [46] | PSRI = (R680 − R500)/(R750) |
| Water Index, related to water content [40] | WI = (R900/R970) |
| Green region NDVI, related to salinity [13] | NDVIgreen = (R550 − R670)/(R550 + R670) |
| Far red region NDVI, related to salinity [13] | NDVIfar red = (R710 − R670)/(R710 + R670) |
| Simple Ratio Vegetation Index, related to salinity [37] | SRVI = (R830/R660) |
| Green and Indigo Ratio, related to salinity [47] | GIR = (R436/R554) |
| Index | F-Fisher | p-Value | Mean | Standard Error | |
|---|---|---|---|---|---|
| LSI | 3440.2 | 0 | Ct | 0.01765a | 0.00045 |
| S1 | 0.03130b | 0.00045 | |||
| S2 | 0.05175c | 0.00045 | |||
| S3 | 0.07829d | 0.00045 | |||
| NDVI | 137.3 | 2.45 × 10−88 | Ct | 0.82469a | 0.00080 |
| S1 | 0.80969b | 0.00080 | |||
| S2 | 0.80345c | 0.00080 | |||
| S3 | 0.80714d | 0.00080 | |||
| NDVI705 | 874.2 | 0 | Ct | 0.45693a | 0.00076 |
| S1 | 0.46466b | 0.00076 | |||
| S2 | 0.47856c | 0.00076 | |||
| S3 | 0.50774d | 0.00076 | |||
| RPI | 686.4 | 0 | Ct | 3.48135a | 0.00864 |
| S1 | 3.54853b | 0.00864 | |||
| S2 | 3.64758c | 0.00864 | |||
| S3 | 3.99075d | 0.00864 | |||
| Chlgreen | 737.2 | 0 | Ct | 2.26632a | 0.00731 |
| S1 | 2.28408a | 0.00731 | |||
| S2 | 2.39855b | 0.00731 | |||
| S3 | 2.69576c | 0.00731 | |||
| Chlred edge | 1024.1 | 0 | Ct | 1.78446a | 0.00583 |
| S1 | 1.84552b | 0.00583 | |||
| S2 | 1.94359c | 0.00583 | |||
| S3 | 2.20687d | 0.00583 | |||
| PSRI | 119.8 | 4.38 × 10−77 | Ct | −0.00536a | 0.00013 |
| S1 | −0.00706b | 0.00013 | |||
| S2 | −0.00552a | 0.00013 | |||
| S3 | −0.00366c | 0.00013 | |||
| WI | 53.4 | 2.03 × 10−34 | Ct | 1.28100a | 0.00036 |
| S1 | 1.27616b | 0.00036 | |||
| S2 | 1.27872c | 0.00036 | |||
| S3 | 1.28220d | 0.00036 | |||
| NDVIgreen | 1160.7 | 0 | Ct | 0.53842a | 0.00108 |
| S1 | 0.50228b | 0.00108 | |||
| S2 | 0.47366c | 0.00108 | |||
| S3 | 0.45415d | 0.00108 | |||
| NDVIfar red | 702.8 | 0 | Ct | 0.68830a | 0.00102 |
| S1 | 0.65912b | 0.00102 | |||
| S2 | 0.63788c | 0.00102 | |||
| S3 | 0.62661d | 0.00102 | |||
| SRVI | 119.7 | 5.35 × 10−77 | Ct | 10.53830a | 0.04345 |
| S1 | 9.64646b | 0.04345 | |||
| S2 | 9.46616c | 0.04345 | |||
| S3 | 10.04068d | 0.04345 | |||
| GIR | 1340.2 | 0 | Ct | 0.35211a | 0.00100 |
| S1 | 0.38783b | 0.00100 | |||
| S2 | 0.41381c | 0.00100 | |||
| S3 | 0.43755d | 0.00100 | |||
| Scores PC1 | 2845.88 | 0 | Ct | −1.0902a | 0.000227 |
| S1 | −0.6474b | 0.000228 | |||
| S2 | 0.0851c | 0.000199 | |||
| S3 | 1.1235d | 0.000212 | |||
| Index | F-Fisher | p-Value | Mean | Standard Error | |
|---|---|---|---|---|---|
| LSI | 1491.57 | 0 | Ct | 0.01845 | 0.00131a |
| S1 | 0.03149 | 0.00127b | |||
| S2 | 0.05250 | 0.00127c | |||
| S3 | 0.08018 | 0.00127d | |||
| Scores PC1 | 1197.02 | 0 | Ct | −1.05383 | 0.05122a |
| S1 | −0.63252 | 0.05122b | |||
| S2 | 0.12681 | 0.05122c | |||
| S3 | 1.16911 | 0.05122d | |||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lara, M.Á.; Diezma, B.; Lleó, L.; Roger, J.M.; Garrido, Y.; Gil, M.I.; Ruiz-Altisent, M. Hyperspectral Imaging to Evaluate the Effect of IrrigationWater Salinity in Lettuce. Appl. Sci. 2016, 6, 412. https://doi.org/10.3390/app6120412
Lara MÁ, Diezma B, Lleó L, Roger JM, Garrido Y, Gil MI, Ruiz-Altisent M. Hyperspectral Imaging to Evaluate the Effect of IrrigationWater Salinity in Lettuce. Applied Sciences. 2016; 6(12):412. https://doi.org/10.3390/app6120412
Chicago/Turabian StyleLara, Miguel Ángel, Belén Diezma, Lourdes Lleó, Jean Michel Roger, Yolanda Garrido, María Isabel Gil, and Margarita Ruiz-Altisent. 2016. "Hyperspectral Imaging to Evaluate the Effect of IrrigationWater Salinity in Lettuce" Applied Sciences 6, no. 12: 412. https://doi.org/10.3390/app6120412






