An Accurate Perception Method for Low Contrast Bright Field Microscopy in Heterogeneous Microenvironments †
Abstract
:1. Introduction
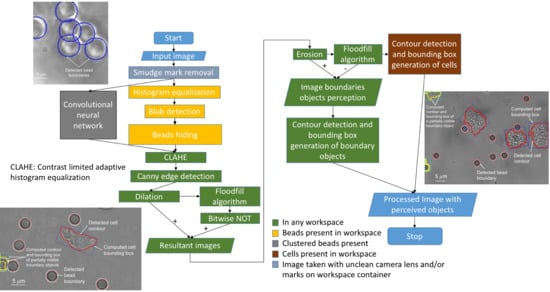
2. Materials and Methods
2.1. Data
2.2. Technical Approach
2.2.1. Smudge Mark Removal
2.2.2. Bead Perception
2.2.3. Cell Perception
3. Results
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ashkin, A. History of Optical Trapping and Manipulation of Small-Neutral Particle, Atoms, and Molecules. IEEE J. Sel. Top. Quantum Electron. 2000, 6, 841–856. [Google Scholar] [CrossRef]
- Gibson, G.M.; Bowman, R.W.; Linnenberger, A.; Dienerowitz, M.; Phillips, D.B.; Carberry, D.M.; Miles, M.J.; Padgett, M.J. A Compact Holographic Optical Tweezers Instrument. Rev. Sci. Instrum. 2012, 83, 113107. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.G.; Chowdhury, S.; Losert, W.; Gupta, S.K. Survey on Indirect Optical Manipulation of Cells, Nucleic Acids, and Motor Proteins. J. Biomed. Opt. 2011, 16, 051302. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.G.; Chowdhury, S.; Gupta, S.K. Optical Tweezers: Autonomous Robots for the Manipulation of Biological Cells. IEEE Robot. Autom. Mag. 2014, 21, 81–88. [Google Scholar] [CrossRef]
- Thakur, A.; Chowdhury, S.; Švec, P.; Wang, C.; Losert, W.; Gupta, S.K. Indirect Pushing based Automated Micromanipulation of Biological Cells using Optical Tweezers. Int. J. Robot. Res. 2014, 33, 1098–1111. [Google Scholar] [CrossRef]
- Banerjee, A.G.; Balijepalli, A.; Gupta, S.K.; LeBrun, T.W. Generating Simplified Trapping Probability Models from Simulation of Optical Tweezers Systems. J. Comput. Inf. Sci. Eng. 2009, 9, 021003. [Google Scholar] [CrossRef]
- Yan, X.; Cheah, C.C.; Pham, Q.-C.; Slotine, J.-J.C. Robotic Manipulation of Micro/Nanoparticles using Optical Tweezers with Velocity Constraints and Stochastic Perturbations. In Proceedings of the IEEE International Conference on Robotics and Automation, Seattle, WA, USA, 26–30 May 2015. [Google Scholar]
- Banerjee, A.G.; Pomerance, A.; Losert, W.; Gupta, S.K. Developing a Stochastic Dynamic Programming Framework for Optical Tweezer-based Automated Particle Transport Operations. IEEE Trans. Autom. Sci. Eng. 2010, 7, 218–227. [Google Scholar] [CrossRef]
- Banerjee, A.G.; Chowdhury, S.; Losert, W.; Gupta, S.K. Real-time Path Planning for Coordinated Transport of Multiple Particles using Optical Tweezers. IEEE Trans. Autom. Sci. Eng. 2012, 9, 669–678. [Google Scholar] [CrossRef]
- Ju, T.; Liu, S.; Yang, J.; Sun, D. Rapidly Exploring Random Tree Algorithm-based Path Planning for Robot-aided Optical Manipulation of Biological Cells. IEEE Trans. Autom. Sci. Eng. 2014, 11, 649–657. [Google Scholar] [CrossRef]
- Chowdhury, S.; Thakur, A.; Švec, P.; Wang, C.; Losert, W.; Gupta, S.K. Automated Manipulation of Biological Cells using Gripper Formations Controlled by Optical Tweezers. IEEE Trans. Autom. Sci. Eng. 2014, 11, 338–347. [Google Scholar] [CrossRef]
- Cheah, C.C.; Li, X.; Yan, X.; Sun, D. Observer-based Optical Manipulation of Biological Cells with Robotic Tweezers. IEEE Trans. Robot. 2014, 30, 68–80. [Google Scholar] [CrossRef]
- Cheah, C.C.; Li, X.; Yan, X.; Sun, D. Simple PD Control Scheme for Robotic Manipulation of Biological Cell. IEEE Trans. Autom. Control 2015, 60, 1427–1432. [Google Scholar] [CrossRef]
- Chen, H.; Sun, D. Swarm-inspired Transportation of Biological Cells using Saturation-controlled Optical Tweezers. In Proceedings of the IEEE International Conference on Robotics and Automation, Seattle, WA, USA, 26–30 May 2015. [Google Scholar]
- Xie, M.; Mills, J.K.; Wang, Y.; Mahmoodi, M.; Sun, D. Automated Translational and Rotational Control of Biological Cells with a Robot-aided Optical Tweezers Manipulation System. IEEE Trans. Autom. Sci. Eng. 2016, 13, 543–551. [Google Scholar] [CrossRef]
- Li, X.; Cheah, C.C.; Ta, Q.M. Cooperative Optical Trapping and Manipulation of Multiple Cells with Robot Tweezers. IEEE Trans. Control Syst. Technol. 2017, 25, 1564–1575. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.; Chen, S.; Wang, Y.; Cheng, S.H.; Sun, D. In Vivo Manipulation of Single Biological Cells With an Optical Tweezers-Based Manipulator and a Disturbance Compensation Controller. IEEE Trans. Robot. 2017, 33, 1200–1212. [Google Scholar] [CrossRef]
- Yang, H.; Li, X.; Liu, Y.; Sun, D. Automated Transportation of Biological Cells for Multiple Processing Steps in Cell Surgery. IEEE Trans. Autom. Sci. Eng. 2017, 14, 1712–1721. [Google Scholar] [CrossRef]
- Li, X.; Cheah, C.C. Stochastic Optical Trapping and Manipulation of Micro Object with Neural-Network Adaptation. IEEE/ASME Trans. Mechatron. 2017, 22, 2633–2642. [Google Scholar] [CrossRef]
- Rajasekaran, K.; Bollavaram, M.; Banerjee, A.G. Toward Automated Formation of Microsphere Arrangements using Multiplexed Optical Tweezers. In Proceedings of the SPIE Optical Trapping and Optical Micromanipulation Conference, San Diego, CA, USA, 28 August–1 September 2016. [Google Scholar]
- Rajasekaran, K.; Samani, E.U.; Stewart, J.; Banerjee, A.G. Imaging-Guided Collision-Free Transport of Multiple Optically Trapped Beads. In Proceedings of the International Conference on Manipulation, Automation, and Robotics at Small Scales, Montréal, QC, Canada, 17–21 July 2017. [Google Scholar]
- Rahman, M.A.; Cheng, J.; Wang, Z.; Ohta, A.T. Cooperative Micromanipulation Using the Independent Actuation of Fifty Microrobots in Parallel. Sci. Rep. 2017, 7, 3278. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.S.S.; Becker, A.; Ou, Y.; Julius, A.A.; Kim, M.J. Swarm Control of Cell-based Microrobots using a Single Global Magnetic Field. In Proceedings of the International Conference on Ubiquitous Robots and Ambient Intelligence, Jeju, Korea, 30 October–2 November 2013. [Google Scholar]
- Felfoul, O.; Martel, S. Assessment of Navigation Control Strategy for Magnetotactic Bacteria in Microchannel: Toward Targeting Solid Tumors. Biomed. Microdevices 2013, 15, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Frutiger, D.R.; Vollmers, K.; Kratochvil, B.E.; Nelson, B.J. Small, Fast, and under Control: Wireless Resonant Magnetic Micro-agents. Int. J. Robot. Res. 2010, 29, 613–636. [Google Scholar] [CrossRef]
- Steager, E.B.; Sakar, M.S.; Magee, C.; Kennedy, M.; Cowley, A.; Kumar, V. Automated Biomanipulation of Single Cells using Magnetic Microrobots. Int. J. Robot. Res. 2013, 32, 346–359. [Google Scholar] [CrossRef]
- Sitti, M.; Ceylan, H.; Hu, W.; Giltinan, J.; Turan, M.; Yim, S.; Diller, E. Biomedical Applications of Untethered Mobile Milli/Microrobots. Proc. IEEE 2015, 103, 205–224. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Jing, W.; Cappelleri, D.J. Towards Independent Control of Multiple Magnetic Mobile Microrobots. Micromachines 2015, 7, 3. [Google Scholar] [CrossRef]
- Chowdhury, S.; Johnson, B.V.; Jing, W.; Cappelleri, D. Designing Local Magnetic Fields and Path Planning for Independent Actuation of Multiple Mobile Microrobots. J. Micro-Bio Robot. 2017, 12, 21–31. [Google Scholar] [CrossRef]
- Banerjee, A.G.; Gupta, S.K. Research in Automated Planning and Control for Micromanipulation. IEEE Trans. Autom. Sci. Eng. 2013, 10, 485–495. [Google Scholar] [CrossRef]
- Chowdhury, S.; Jing, W.; Cappelleri, D. Controlling Multiple Microrobots: Recent Progress and Future Challenges. J. Micro-Bio Robot. 2015, 10, 1–11. [Google Scholar] [CrossRef]
- Chen, X.-Z.; Hoop, M.; Mushtaq, F.; Siringil, E.; Hu, C.; Nelson, B.J.; Pane, S. Recent Developments in Magnetically Driven Micro- and Nanorobots. Appl. Mater. Today 2017, 9, 37–48. [Google Scholar] [CrossRef]
- Peng, T.; Balijepalli, A.; Gupta, S.K.; LeBrun, T. Algorithms for On-line Monitoring of Micro Spheres in an Optical Tweezers-based Assembly Cell. J. Comput. Inf. Sci. Eng. 2007, 7, 330–338. [Google Scholar] [CrossRef]
- Ali, R.; Gooding, M.; Szilágyi, T.; Vojnovic, B.; Christlie, M.; Brady, M. Automatic Segmentation of Adherent Biological Cell Boundaries and Nuclei from Brightfield Microscopy Images. Mach. Vis. Appl. 2012, 23, 607–621. [Google Scholar] [CrossRef]
- Mohamadlou, H.; Shope, J.C.; Flann, N.S. Maximizing Kolmogorov Complexity for Accurate and Robust Bright Field Cell Segmentation. BMC Bioinf. 2014, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Cenev, Z.; Venäläinen, J.; Sariola, V.; Zhou, Q. Object Tracking in Robotic Micromanipulation by Supervised Ensemble Learning Classifier. In Proceedings of the International Conference on Manipulation, Automation and Robotics at Small Scales, Paris, France, 18–22 July 2016. [Google Scholar]
- Buggenthin, F.; Marr, C.; Schwarzfischer, M.; Hoppe, P.S.; Hilsenbeck, O.; Schroeder, T.; Theis, F.J. An Automatic Method for Robust and Fast Cell Detection in Bright Field Images from High-throughput Microscopy. BMC Bioinform. 2013, 14, 297. [Google Scholar] [CrossRef] [PubMed]
- Hajiyavand, A.M.; Saadat, M.; Bedi, A.-P.S. Polar Body Detection for ICSI Cell Manipulation. In Proceedings of the International Conference on Manipulation, Automation and Robotics at Small Scales, Paris, France, 18–22 July 2016. [Google Scholar]
- Bollavaram, M.; Sane, P.; Chowdhury, S.; Gupta, S.K.; Banerjee, A.G. Automated Detection of Live Cells and Microspheres in Low Contrast Bright Field Microscopy. In Proceedings of the International Conference on Manipulation, Automation and Robotics at Small Scales, Paris, France, 18–22 July 2016. [Google Scholar]
- Gu, J.; Ramamoorthi, R.; Belhumeur, P.; Nayar, S. Removing Image Artifacts due to Dirty Camera Lenses and Thin Occluders. ACM Trans. Graph. 2009, 28, 144. [Google Scholar] [CrossRef]
- Suzuki, S.; Abe, K. Topological structural analysis of digitized binary images by border following. Comput. Graph. Image Process. 1985, 30, 32–46. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Pizer, S.M.; Amburn, E.P.; Austin, J.D.; Cromartie, R.; Geselowitz, A.; Greer, T.; Romeny, B.H.; Zimmerman, J.B.; Zuiderveld, K. Adaptive Histogram Equalization and its Variations. Comput. Graph. Image Process. 1987, 39, 355–368. [Google Scholar] [CrossRef]
- Canny, J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 6, 679–698. [Google Scholar] [CrossRef]
- Toussaint, G.T. Solving Geometric Problems with the Rotating Calipers. In Proceedings of the IEEE Mediterranean Electrotechnical Conference, Athens, Greece, 24–26 May 1983. [Google Scholar]
- Keras. Available online: https://github.com/fchollet/keras (accessed on 1 October 2017).
- Nister, D.; Stewenius, H. Linear Time Maximally Stable Extremal Regions. In Proceedings of the European Conference on Computer Vision, Marseille, France, 12–18 October 2008. [Google Scholar]
- Bay, H.; Ess, A.; Tuytelaars, T.; Gool, L.V. Speeded-Up Robust Features (SURF). Comput. Vis. Image Underst. 2008, 110, 346–359. [Google Scholar] [CrossRef]
- Fang, M.; Yue, G.X.; Yu, Q.C. The Study on an Application of Otsu Method in Canny Operator. In Proceedings of the International Symposium on Information Processing, Huangshan, China, 21–23 August 2009. [Google Scholar]
- Guo, W.; Manohar, K.; Brunton, S.; Banerjee, A.G. Sparse-TDA: Sparse Realization of Topological Data Analysis for Multi-Way Classification. arXiv, 2017; arXiv:1701.03212. [Google Scholar]


























© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajasekaran, K.; Samani, E.; Bollavaram, M.; Stewart, J.; Banerjee, A.G. An Accurate Perception Method for Low Contrast Bright Field Microscopy in Heterogeneous Microenvironments. Appl. Sci. 2017, 7, 1327. https://doi.org/10.3390/app7121327
Rajasekaran K, Samani E, Bollavaram M, Stewart J, Banerjee AG. An Accurate Perception Method for Low Contrast Bright Field Microscopy in Heterogeneous Microenvironments. Applied Sciences. 2017; 7(12):1327. https://doi.org/10.3390/app7121327
Chicago/Turabian StyleRajasekaran, Keshav, Ekta Samani, Manasa Bollavaram, John Stewart, and Ashis G. Banerjee. 2017. "An Accurate Perception Method for Low Contrast Bright Field Microscopy in Heterogeneous Microenvironments" Applied Sciences 7, no. 12: 1327. https://doi.org/10.3390/app7121327