Automatic Definition of an Anatomic Field of View for Volumetric Cardiac Motion Estimation at High Temporal Resolution
Abstract
:1. Introduction
2. Methods
2.1. Anatomical Relevant Space
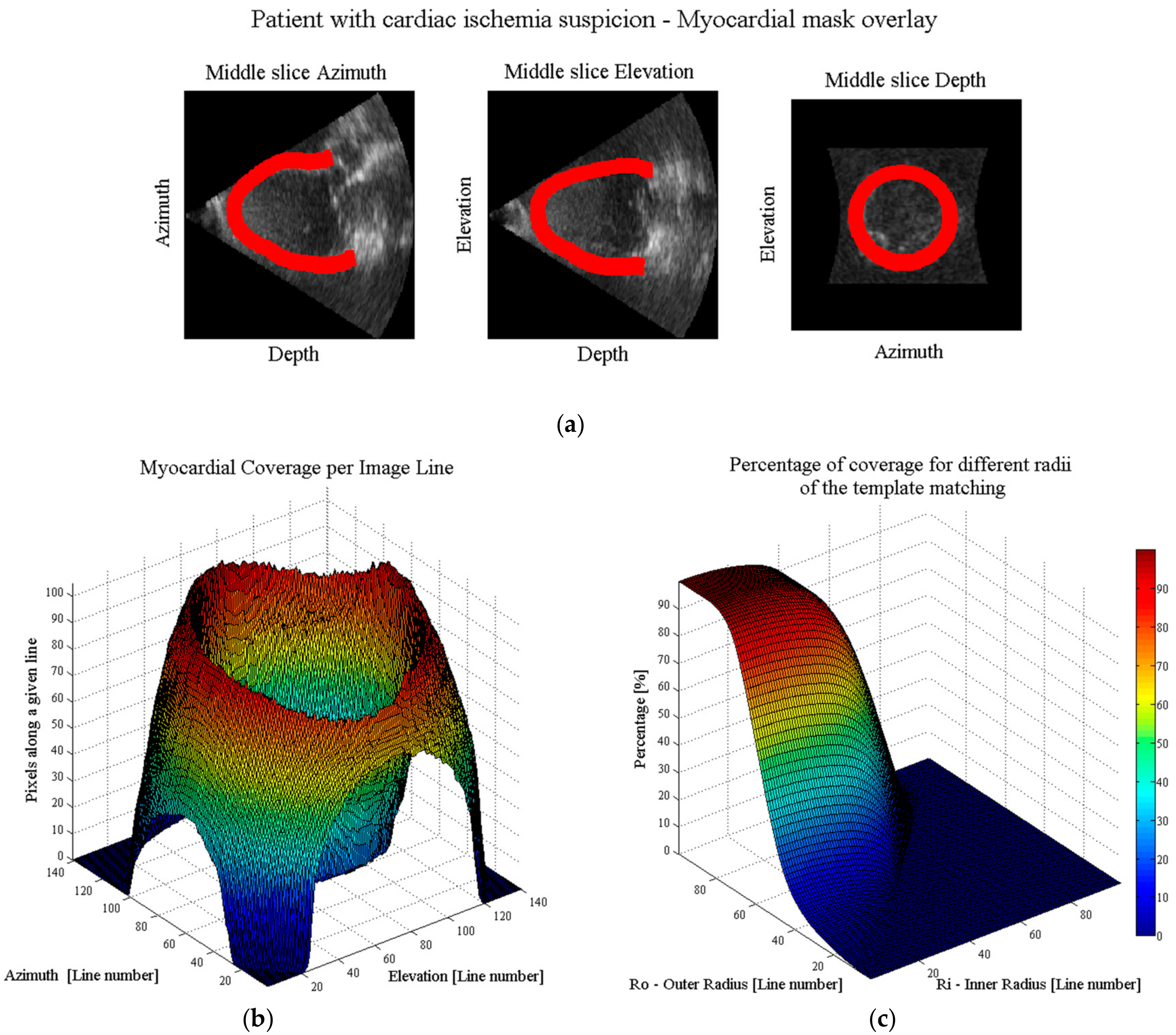
- Automatic real-time segmentation of myocardial boundariesA fully automatic real-time segmentation of the left ventricular myocardium in a volumetric ultrasound recording was performed using the B-spline Explicit Active Surfaces (BEAS) framework [18,19]. More specifically, BEAS uses two explicit functions, one to represent the endocardial surface and another to represent the myocardial thickness. This allows to fully characterize the endo- and epicardial surfaces. These surfaces can then be used to define a binary mask identifying voxels belonging to the myocardium only.
- Coverage functionUsing these binary images, a “coverage function” was defined as follows. First, based on the ray-tracing principle, the path of a given scanline within the volumetric image volume can be traced. The pixels belonging to that scanline are compared with the binary mask, in order to compute the percentage of pixels of the given scanline belonging to the myocardium. Finally, this procedure is repeated for all scan lines in the original pyramidal volume leading to a “coverage function”.
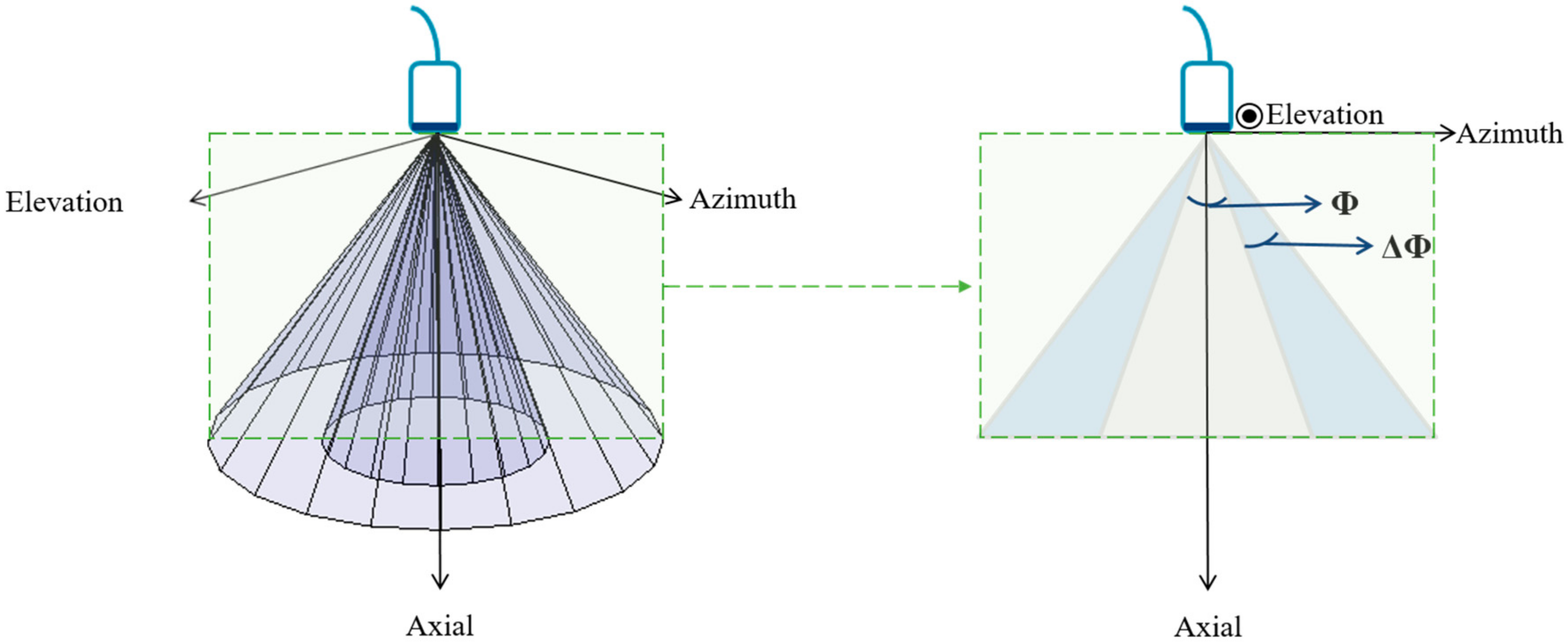
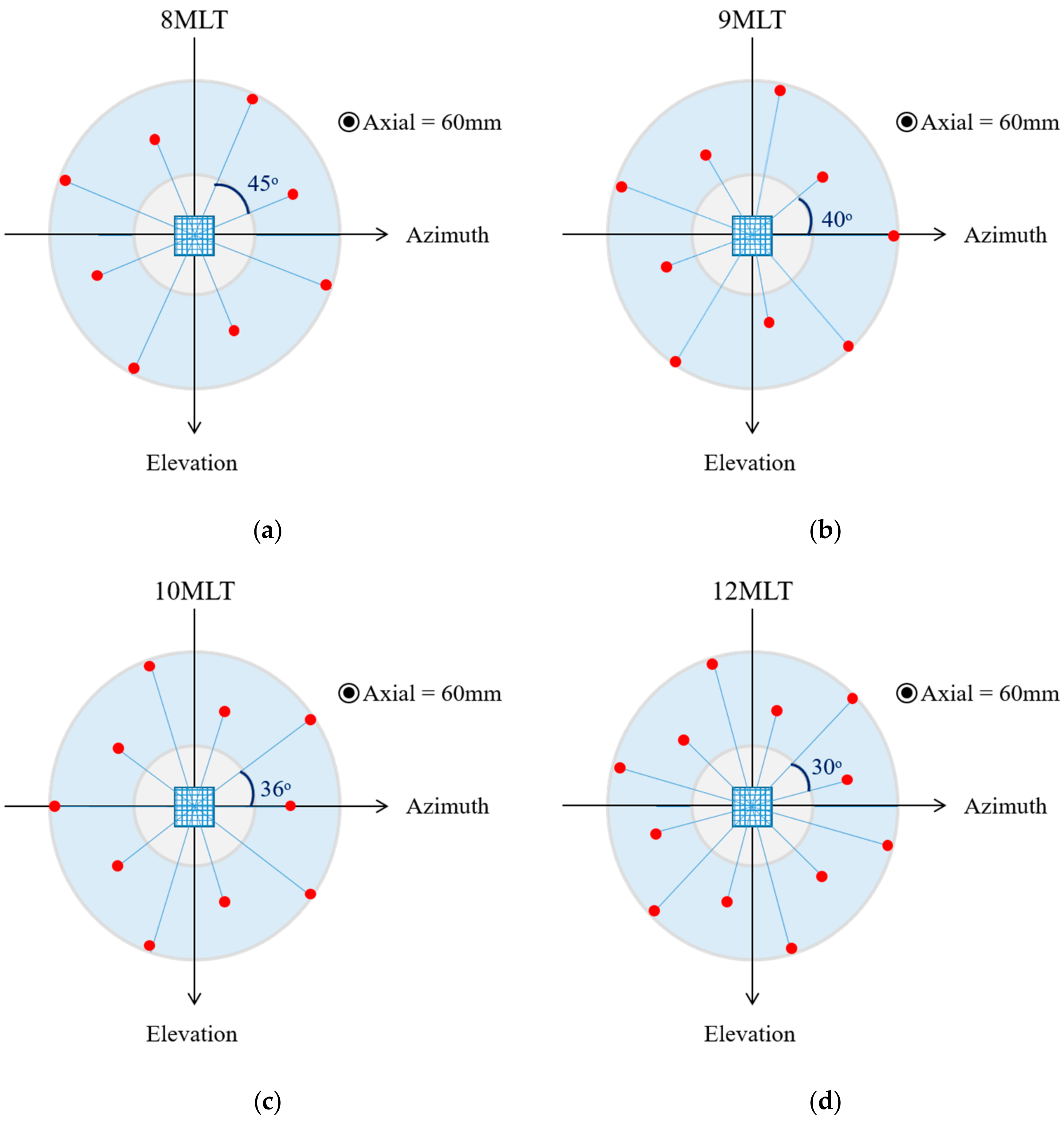
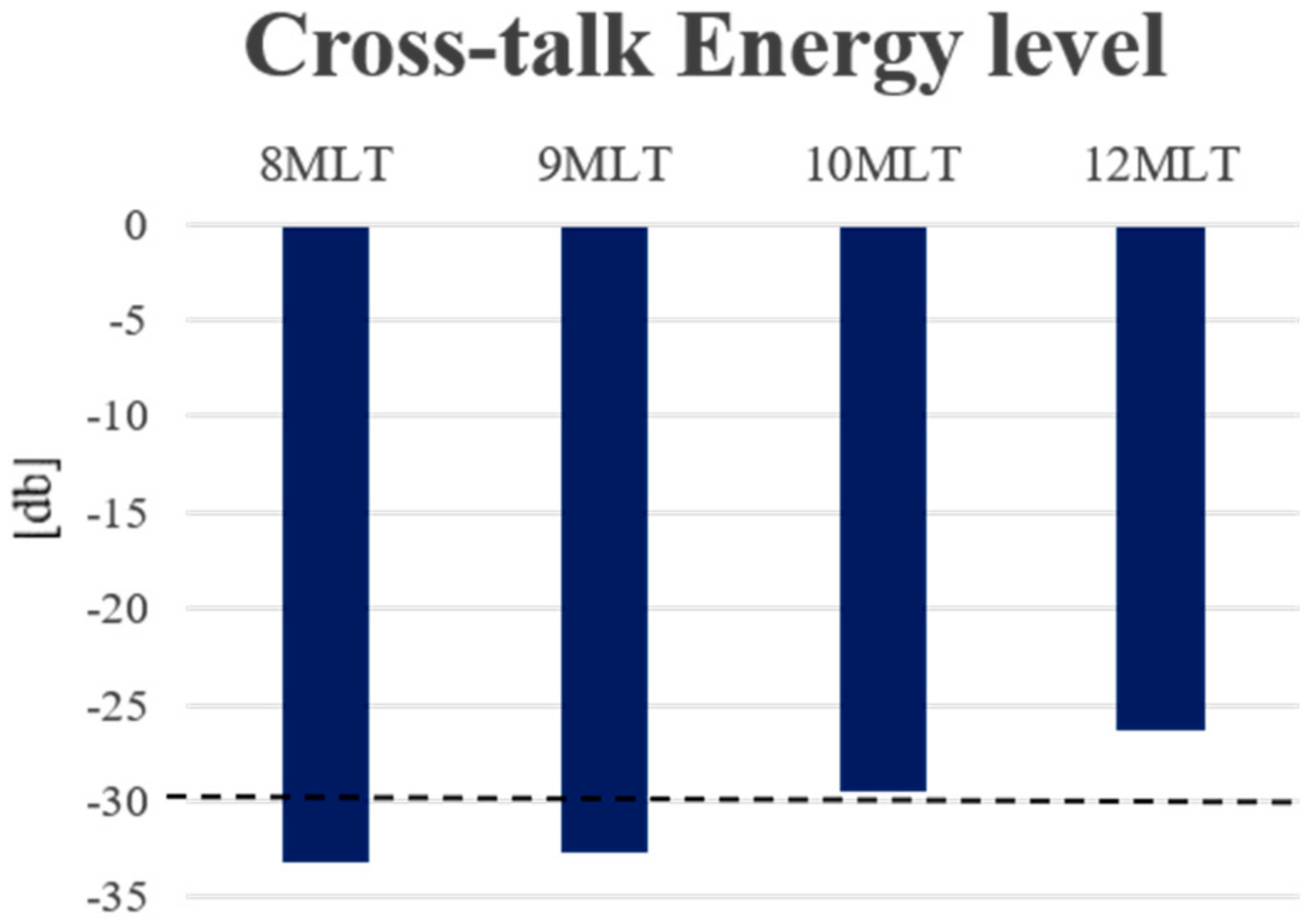
- Ring-shaped template matchingTo find a spatially continuous FOV that covers a given percentage of the total amount of myocardium (i.e., prospectively defined by the user as “T”), a ring-shaped template matching was used. This shape was chosen as an approximation of the left ventricular geometry when looking down from the apex, i.e., when the transducer is placed in an apical position. In 3D, this FOV therefore defines a hollowed cone, Figure 1 (left). We express the amount of myocardial coverage T as a function of the inner radius of the ring template () and its thickness (). In order to effectively gain frame rate, a compromise has to be made between the amount of myocardial coverage, i.e., T, and the extent of the FOV. From all and combinations that provide T myocardial coverage, the one with minimal was chosen, as this would keep the volume to be scanned minimal. In this way, it is ensured that the desired T coverage is obtained using the least amount of lines possible (i.e., at the highest frame rate). In turn, these radii are used to determine the parameters (opening angle, Φ, and thickness, dΦ) for a conical scan, as represented in Figure 1 (right). These radii give directly the inner and outer image lines that define the inner and outer surface of the cone. Then, using the angular inter-beam spacing, the line numbers can be converted to the respective angles.
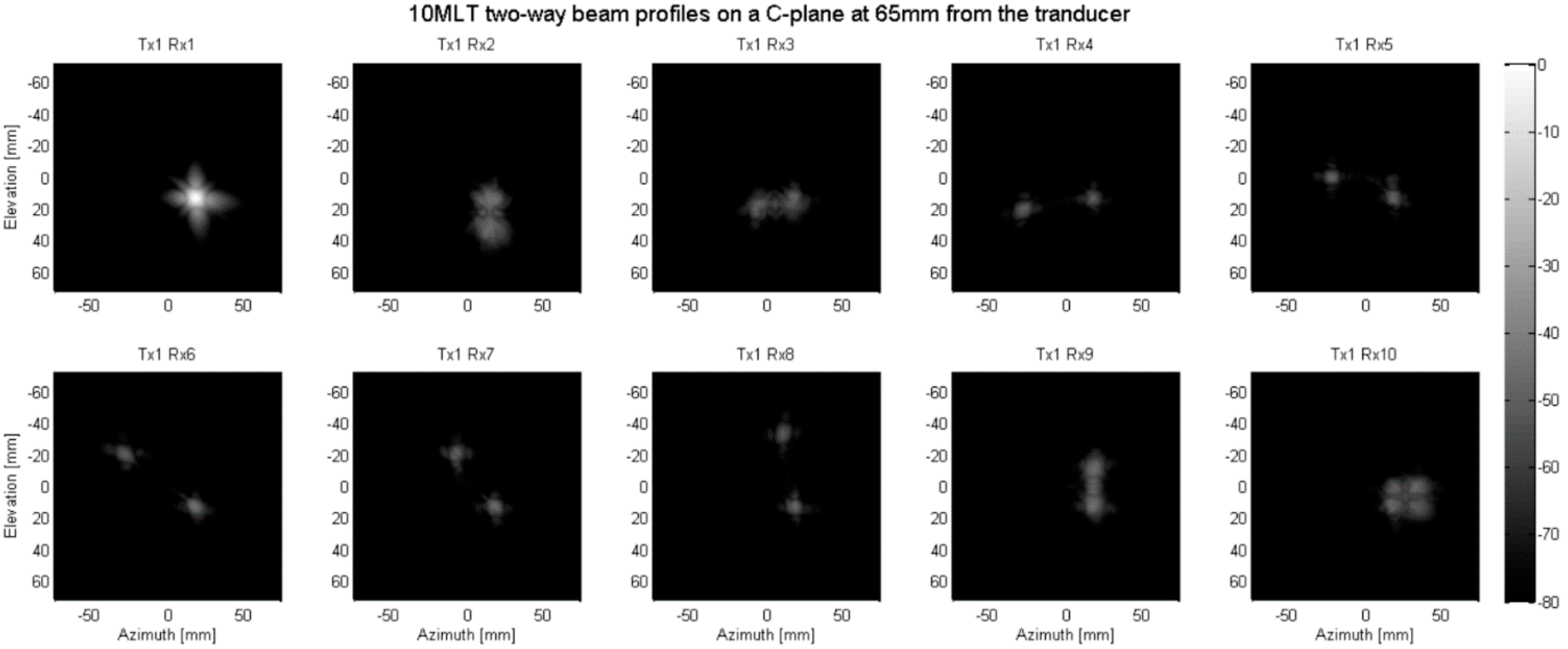
2.2. Parallelized Scan Sequence
3. Experiments
4. Results
5. Discussion
- A pyramidal volume is acquired at a conventional frame rate (i.e., ~30 Hz).
- The coverage function and the ring-shaped template matching are applied to define an anatomical conically shaped FOV.
- Based on (iii), a fast scanning sequence is automatically selected using a LUT giving the best combination of transmit and receive parallelization (i.e., MLT-MLA) to scan the detected region-of-interest.
- The anatomical relevant space is scanned at high spatiotemporal resolution for subsequent motion analysis.
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lang, R.M.; Badano, L.P.; Mor-Avi, V.; Afilalo, J.; Armstrong, A.; Ernande, L.; Flachskampf, F.A.; Foster, E.; Goldstein, S.A.; Kuznetsova, T.; et al. Recommendations for cardiac chamber quantification by echocardiography in adults: An update from the American society of echocardiography and the European association of cardiovascular imaging. Eur. Heart J. Cardiovasc. Imaging 2015, 16, 233–271. [Google Scholar] [CrossRef] [PubMed]
- Jasaityte, R.; Heyde, B.; D’hooge, J. Current state of three-dimensional myocardial strain estimation using echocardiography. J. Am. Soc. Echocardiogr. 2013, 26, 15–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’hooge, J.; Konofagou, E.; Jamal, F.; Heimdal, A.; Barrios, L.; Bijnens, B.; Thoen, J.; Van de Werf, F.; Sutherland, G.R.; Suetens, P. Two-dimensional ultrasonic strain rate measurement of the human heart in vivo. Ultrason. Ferroelectr. Freq. Control. IEEE Trans. 2002, 49, 281–286. [Google Scholar] [CrossRef]
- Kanai, H.; Koiwa, Y. Myocardial rapid velocity distribution. Ultrasound Med. Biol. 2001, 27, 481–498. [Google Scholar] [CrossRef]
- Pernot, M.; Fujikura, K.; Fung-Kee-Fung, S.D.; Konofagou, E.E. ECG-gated, mechanical and electromechanical wave imaging of cardiovascular tissues in vivo. Ultrasound Med. Biol. 2007, 33, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Shattuck, D.P.; Weinshenker, M.D.; Smith, S.W.; von Ramm, O.T. Explososcan: A parallel processing technique for high speed ultrasound imaging with linear phased arrays. J. Acoust. Soc. Am. 1984, 75, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Sarvazyan, A.P.; Rudenko, O.V.; Swanson, S.D.; Fowlkes, J.B.; Emelianov, S.Y. Shear Wave Elasticity Imaging: A new ultrasonic technology of medical diagnostic. Ultrasound Med. Biol. 1998, 24, 1419–1435. [Google Scholar] [CrossRef]
- Provost, J.; Papadacci, C.; Arango, J.E.; Imbault, M.; Fink, M.; Gennisson, J.-L.; Tanter, M.; Pernot, M. 3D ultrafast ultrasound imaging in vivo. Phys. Med. Biol. 2014, 59, L1–L13. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.F.; Trahey, G.E. A fundamental limit on the performance of correlation based phase correction and flow estimation techniques. In Proceedings of the IEEE Ultrasonics Symposium, Montréal, QC, Canada, 23–27 August 2004. [Google Scholar]
- Prieur, F.; Dénarié, B.; Austeng, A.; Torp, H. Multi-Line Transmission in Medical Imaging Using the Second-Harmonic Signal. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2013, 60, 2682–2692. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Gao, H.; D’hooge, J. Multi-transmit beamforming for fast cardiac imaging-a simulation study. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2013, 60, 1719–1731. [Google Scholar] [CrossRef] [PubMed]
- Demi, L.; Verweij, M.D.; Van Dongen, K.W. Parallel transmit beamforming using orthogonal frequency division multiplexing applied to harmonic imaging—A feasibility study. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2012, 59, 2439–2447. [Google Scholar] [CrossRef] [PubMed]
- Demi, L.; Ramalli, A.; Giannini, G.; Mischi, M. In Vitro and in Vivo tissue harmonic images obtained with parallel transmit beamforming by means of orthogonal frequency division multiplexing. IEEE Ultrason. Symp. Proc. 2015, 62, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Hergum, T.; Bjastad, T.; Kristoffersen, K.; Torp, H. Parallel Beamforming Using Synthetic Transmit Beams. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2007, 54, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.; Tong, L.; Ortega, A.; Løvstakken, L.; Samset, E.; D’hooge, J. Acoustic Output of Multi-Line Transmit Beamforming for Fast Cardiac Imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2015, 62, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Ortega, A.; Gao, H.; D’hooge, J. Fast three-dimensional ultrasound cardiac imaging using multi-transmit beamforming: A simulation study. IEEE Ultrason. Symp. Proc. 2013, 60, 1456–1459. [Google Scholar]
- Ortega, A.; Provost, J.; Tong, L.; Santos, P.; Heyde, B.; Pernot, M.; D’hooge, J. A comparison of the performance of different multi-line transmit setups for fast volumetric cardiac ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2016, 63, 1. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, D.; Dietenbeck, T.; Schaerer, J.; D’hooge, J.; Friboulet, D.; Bernard, O. B-spline explicit active surfaces: An efficient framework for real-time 3-D region-based segmentation. IEEE Trans. Image Process. 2012, 21, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, J.; Barbosa, D.; Heyde, B.; Schnell, F.; Rösner, A.; Claus, P.; D’hooge, J. Left Ventricular Myocardial Segmentation in 3-D Ultrasound recordings: Effect of Different Endocardial and Epicardial Coupling Strategies. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2017, 64, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Bruyneel, T.; Ortega, A.; Tong, L.; D’hooge, J. A GPU-based implementation of the spatial impulse response method for fast calculation of linear sound fields and pulse-echo responses of array transducers. Proceedings of IEEE Ultrasonic Symposium, Prague, Czech Republic, 21–25 July 2013. [Google Scholar]
- Rademakers, F.; Engvall, J.; Edvardsen, T.; Monaghan, M.; Sicari, R.; Nagel, E.; Zamorano, J.; Ukkonen, H.; Ebbers, T.; Di Bello, V.; et al. Determining optimal noninvasive parameters for the prediction of left ventricular remodeling in chronic ischemic patients. J. Scand. Cardiovasc. 2013, 47, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Ramalli, A.; Jasaityte, R.; Tortoli, P.; D’hooge, J. Multi-transmit beamforming for fast cardiac imaging—Experimental validation and in vivo application. IEEE Trans. Med. Imag. 2014, 33, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Ramalli, A.; Dallai, A.; Boni, E.; Bassi, L.; Meacci, V.; Giovannetti, M.; Tong, L.; D’hooge, J.; Tortoli, P. Multi transmit beams for fast cardiac imaging towards clinical routine. In Proceedings of the IEEE International Ultrasonics Symposium (IUS), Tours, France, 18–21 September 2016. [Google Scholar]
- Barbosa, D.; Dietenbeck, T.; Heyde, B.; Houle, H.; Friboulet, D.; D’hooge, J.; Bernard, O. Fast and fully automatic 3D echocardiographic segmentation using B-spline explicit active surfaces: Feasibility study and validation in a clinical setting. Ultrasound Med Biol. 2013, 39, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Orderud, F.; Rabben, S.I. Real-time 3D segmentation of the left ventricle using deformable subdivision surfaces. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Anchorage, AK, USA, 23–28 June 2008. [Google Scholar]
- Jurie, F.; Dhome, M. Real Time Robust Template Matching. In Proceedings of the 13th British Machine Vision Conference, University of Cardiff, Cardiff, Wales, 2–5 September 2002; pp. 1–10. [Google Scholar]
- Savord, B.J. Beamforming Methods and Apparatus for Three-Dimensional Ultrasound Imaging Using Two-Dimensional Transducer Array. U.S. Patent 6013032A, 11 January 2000. [Google Scholar]
- Santos, P.; Haugen, G.; Løvstakken, L.; Samset, E.; D’hooge, J. Diverging Wave Volumetric Imaging Using Sub-Aperture Beamforming. IEEE Trans. Med. Imag. 2016, 63, 2114–2124. [Google Scholar]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega, A.; Pedrosa, J.; Heyde, B.; Tong, L.; D’hooge, J. Automatic Definition of an Anatomic Field of View for Volumetric Cardiac Motion Estimation at High Temporal Resolution. Appl. Sci. 2017, 7, 752. https://doi.org/10.3390/app7070752
Ortega A, Pedrosa J, Heyde B, Tong L, D’hooge J. Automatic Definition of an Anatomic Field of View for Volumetric Cardiac Motion Estimation at High Temporal Resolution. Applied Sciences. 2017; 7(7):752. https://doi.org/10.3390/app7070752
Chicago/Turabian StyleOrtega, Alejandra, João Pedrosa, Brecht Heyde, Ling Tong, and Jan D’hooge. 2017. "Automatic Definition of an Anatomic Field of View for Volumetric Cardiac Motion Estimation at High Temporal Resolution" Applied Sciences 7, no. 7: 752. https://doi.org/10.3390/app7070752





