Morphological and Molecular Basis of Cytoplasmic Dilation and Swelling in Cortical Migrating Neurons
Abstract
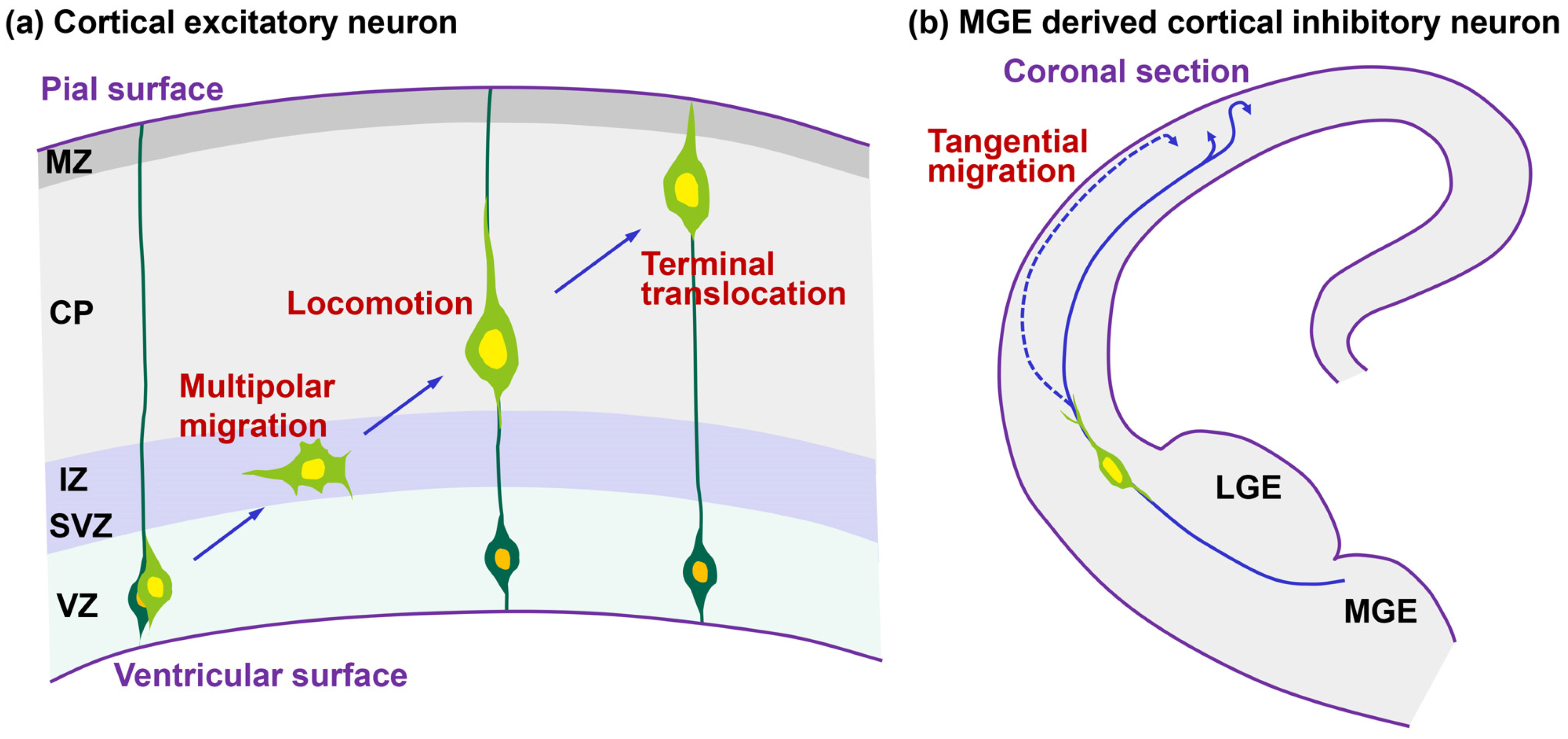
:1. Introduction
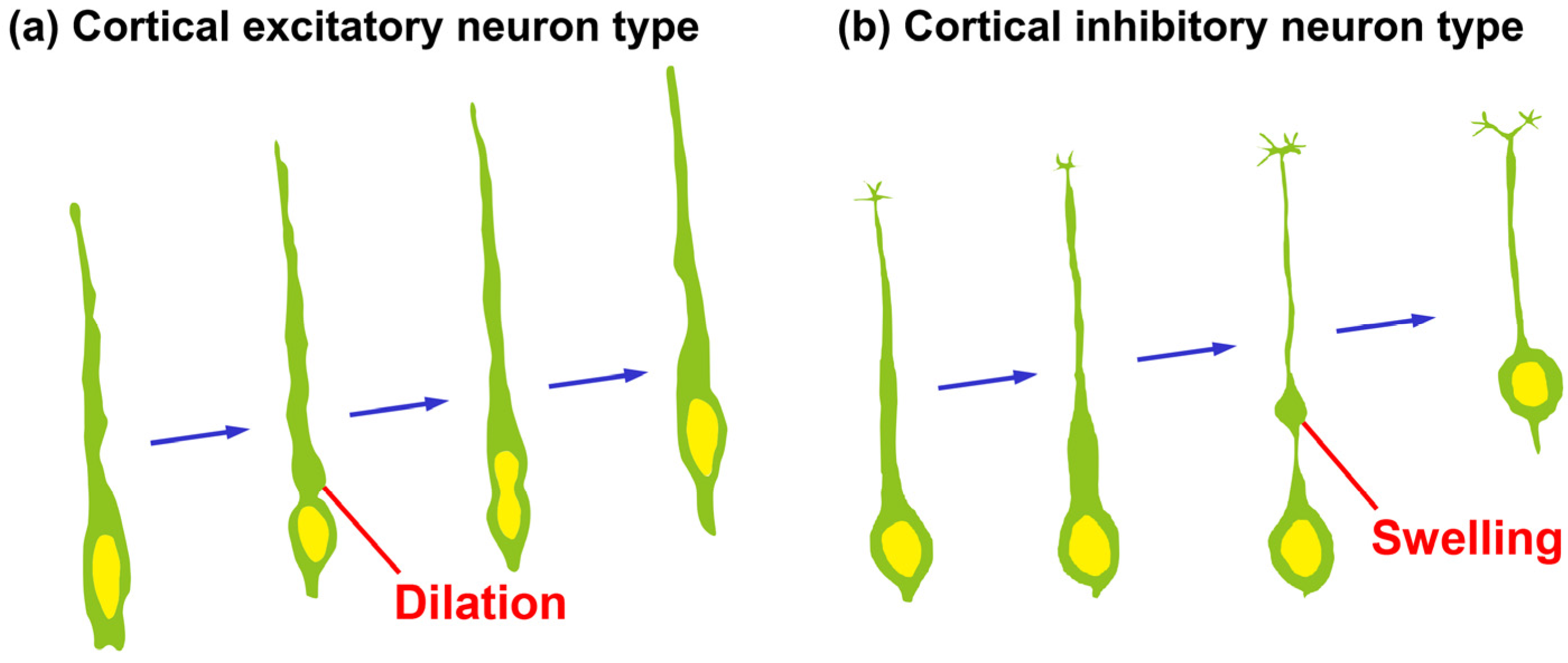
2. Dilation/Swelling Formation
2.1. Morphological Features of Dilation and Swelling in Migrating Neurons
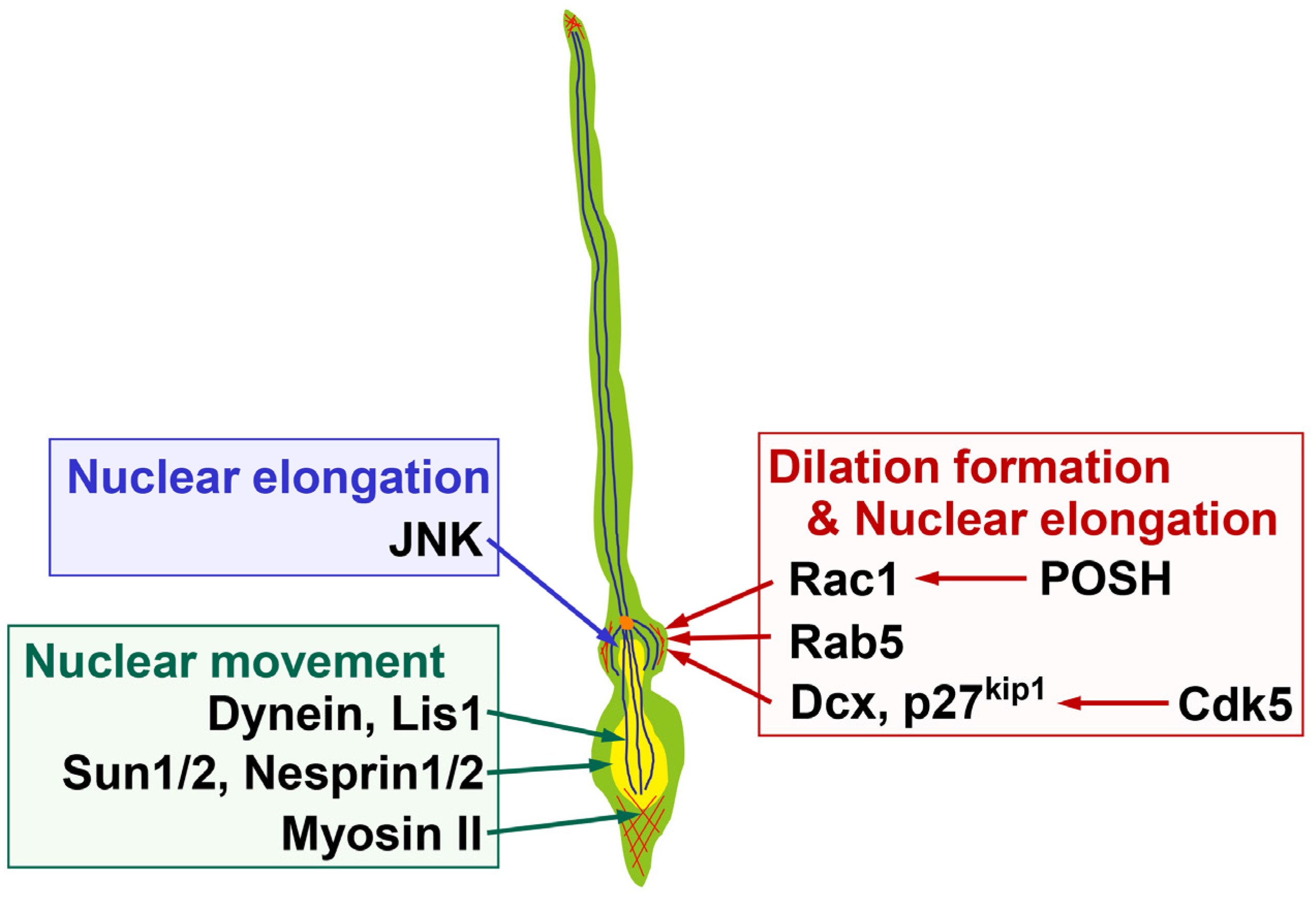
2.2. Molecular Mechanisms Underlying Dilation and Swelling Formation
3. Nuclear Deformation and Movement
3.1. Nuclear Deformation during the Locomotion
3.2. Molecules Regulating Nuclear Elongation
3.3. The Forward Movement of the Nucleus
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gleeson, J.G.; Walsh, C.A. Neuronal migration disorders: From genetic diseases to developmental mechanisms. Trends Neurosci. 2000, 23, 352–359. [Google Scholar] [CrossRef]
- Kawauchi, T.; Hoshino, M. Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Dev. Neurosci. 2008, 30, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Ayala, R.; Shu, T.; Tsai, L.H. Trekking across the brain: The journey of neuronal migration. Cell 2007, 128, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Heng, J.I.; Chariot, A.; Nguyen, L. Molecular layers underlying cytoskeletal remodelling during cortical development. Trends Neurosci. 2010, 33, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.A. Molecules and mechanisms that regulate multipolar migration in the intermediate zone. Front Cell. Neurosci. 2014, 8, 386. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, T. Cellullar insights into cerebral cortical development: Focusing on the locomotion mode of neuronal migration. Front Cell. Neurosci. 2015, 9, 394. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Anton, E.S. Decision making during interneuron migration in the developing cerebral cortex. Trends Cell Biol. 2014, 24, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Marin, O.; Valiente, M.; Ge, X.; Tsai, L.H. Guiding neuronal cell migrations. Cold Spring Harb. Perspect. Biol. 2010, 2, a001834. [Google Scholar] [CrossRef] [PubMed]
- Metin, C.; Baudoin, J.P.; Rakic, S.; Parnavelas, J.G. Cell and molecular mechanisms involved in the migration of cortical interneurons. Eur. J. Neurosci. 2006, 23, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Rakic, P. A century of progress in corticoneurogenesis: From silver impregnation to genetic engineering. Cereb. Cortex 2006, 16, i3–i17. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.V.; Sekine, K.; Chihama, K.; Nakajima, K.; Hoshino, M.; Nabeshima, Y.; Kawauchi, T. Dissecting the factors involved in the locomotion mode of neuronal migration in the developing cerebral cortex. J. Biol. Chem. 2010, 285, 5878–5887. [Google Scholar] [CrossRef] [PubMed]
- Schaar, B.T.; McConnell, S.K. Cytoskeletal coordination during neuronal migration. Proc. Natl. Acad. Sci. USA 2005, 102, 13652–13657. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.V.; Shikanai, M.; Hoshino, M.; Ohshima, T.; Nabeshima, Y.; Mizutani, K.; Nagata, K.; Nakajima, K.; Kawauchi, T. Cdk5 and its substrates, Dcx and p27kip1, regulate cytoplasmic dilation formation and nuclear elongation in migrating neurons. Development 2014, 141, 3540–3550. [Google Scholar] [CrossRef] [PubMed]
- Nichols, A.J.; Carney, L.H.; Olson, E.C. Comparison of slow and fast neocortical neuron migration using a new in vitro model. BMC Neurosci. 2008, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Asada, N.; Sanada, K. LKB1-mediated spatial control of GSK3beta and adenomatous polyposis coli contributes to centrosomal forward movement and neuronal migration in the developing neocortex. J. Neurosci. 2010, 30, 8852–8865. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, T.; Sekine, K.; Shikanai, M.; Chihama, K.; Tomita, K.; Kubo, K.; Nakajima, K.; Nabeshima, Y.; Hoshino, M. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron 2010, 67, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Bellion, A.; Baudoin, J.P.; Alvarez, C.; Bornens, M.; Metin, C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: Forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 2005, 25, 5691–5699. [Google Scholar] [CrossRef] [PubMed]
- Anton, E.S.; Marchionni, M.A.; Lee, K.F.; Rakic, P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development 1997, 124, 3501–3510. [Google Scholar] [PubMed]
- Chae, T.; Kwon, Y.T.; Bronson, R.; Dikkes, P.; Li, E.; Tsai, L.H. Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron 1997, 18, 29–42. [Google Scholar] [CrossRef]
- Gilmore, E.C.; Ohshima, T.; Goffinet, A.M.; Kulkarni, A.B.; Herrup, K. Cyclin-dependent kinase 5-deficient mice demonstrate novel developmental arrest in cerebral cortex. J. Neurosci. 1998, 18, 6370–6377. [Google Scholar] [PubMed]
- Kawauchi, T.; Chihama, K.; Nabeshima, Y.; Hoshino, M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003, 22, 4190–4201. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, T.; Chihama, K.; Nabeshima, Y.; Hoshino, M. Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat. Cell Biol. 2006, 8, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, T.; Hirasawa, M.; Tabata, H.; Mutoh, T.; Adachi, T.; Suzuki, H.; Saruta, K.; Iwasato, T.; Itohara, S.; Hashimoto, M.; et al. Cdk5 is required for multipolar-to-bipolar transition during radial neuronal migration and proper dendrite development of pyramidal neurons in the cerebral cortex. Development 2007, 134, 2273–2282. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, G.; An, L.; Fan, Y.; Cheng, X.; Li, X.; Yin, Y.; Cong, R.; Chen, S.; Zhao, S. Fyn regulates multipolar-bipolar transition and neurite morphogenesis of migrating neurons in the developing neocortex. Neuroscience 2017, 352, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.W.; Bremner, K.H.; Vallee, R.B. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007, 10, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Godin, J.D.; Thomas, N.; Laguesse, S.; Malinouskaya, L.; Close, P.; Malaise, O.; Purnelle, A.; Raineteau, O.; Campbell, K.; Fero, M.; et al. p27Kip1 is a microtubule-associated protein that promotes microtubule polymerization during neuron migration. Dev. Cell 2012, 23, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Shieh, J.C.; Schaar, B.T.; Srinivasan, K.; Brodsky, F.M.; McConnell, S.K. Endocytosis regulates cell soma translocation and the distribution of adhesion proteins in migrating neurons. PLoS ONE 2011, 6, e17802. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Sun, Y.; Zhang, F.; Zhu, Y.; Shi, L.; Li, H.; Xu, Z. POSH localizes activated Rac1 to control the formation of cytoplasmic dilation of the leading process and neuronal migration. Cell Rep. 2012, 2, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Solecki, D.J.; Trivedi, N.; Govek, E.E.; Kerekes, R.A.; Gleason, S.S.; Hatten, M.E. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron 2009, 63, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, T.; Chihama, K.; Nishimura, Y.V.; Nabeshima, Y.; Hoshino, M. MAP1B phosphorylation is differentially regulated by Cdk5/p35, Cdk5/p25, and JNK. Biochem. Biophys. Res. Commun. 2005, 331, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Takei, Y.; Teng, J.; Harada, A.; Hirokawa, N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J. Cell Biol. 2000, 150, 989–1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.; Ramos, R.L.; Ackman, J.B.; Thomas, A.M.; Lee, R.V.; LoTurco, J.J. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat. Neurosci. 2003, 6, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.T.; Kerjan, G.; Bielas, S.L.; Lee, J.E.; Fenstermaker, A.G.; Novarino, G.; Gleeson, J.G. Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microRNA dysregulation. Neuron 2014, 82, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
- Pramparo, T.; Youn, Y.H.; Yingling, J.; Hirotsune, S.; Wynshaw-Boris, A. Novel embryonic neuronal migration and proliferation defects in Dcx mutant mice are exacerbated by Lis1 reduction. J. Neurosci. 2010, 30, 3002–3012. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, M.; Prokscha, A.; Ungewickell, E.; Eichele, G. Doublecortin association with actin filaments is regulated by neurabin II. J. Biol. Chem. 2005, 280, 11361–11368. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Brown, K.J.; Yap, C.C.; Winckler, B.; Jaiswal, J.K.; Liu, J.S. Doublecortin (Dcx) family proteins regulate filamentous actin structure in developing neurons. J. Neurosci. 2013, 33, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Yap, C.C.; Vakulenko, M.; Kruczek, K.; Motamedi, B.; Digilio, L.; Liu, J.S.; Winckler, B. Doublecortin (DCX) mediates endocytosis of neurofascin independently of microtubule binding. J. Neurosci. 2012, 32, 7439–7453. [Google Scholar] [CrossRef] [PubMed]
- Friocourt, G.; Chafey, P.; Billuart, P.; Koulakoff, A.; Vinet, M.C.; Schaar, B.T.; McConnell, S.K.; Francis, F.; Chelly, J. Doublecortin interacts with mu subunits of clathrin adaptor complexes in the developing nervous system. Mol. Cell. Neurosci. 2001, 18, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Serneo, F.F.; Tseng, H.C.; Kulkarni, A.B.; Tsai, L.H.; Gleeson, J.G. Cdk5 phosphorylation of doublecortin ser297 regulates its effect on neuronal migration. Neuron 2004, 41, 215–227. [Google Scholar] [CrossRef]
- Kawauchi, T. Cdk5 regulates multiple cellular events in neural development, function and disease. Dev. Growth Differ. 2014, 56, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; Roberts, J.M. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999, 13, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, T.; Aoki, Y.; Eksioglu, Y.Z.; Takahashi, T.; Bhide, P.G.; Reeves, S.A.; Caviness, V.S., Jr. Overexpression of p27Kip1 lengthens the G1 phase in a mouse model that targets inducible gene expression to central nervous system progenitor cells. Proc. Natl. Acad. Sci. USA 2001, 98, 6435–6440. [Google Scholar] [CrossRef] [PubMed]
- Tarui, T.; Takahashi, T.; Nowakowski, R.S.; Hayes, N.L.; Bhide, P.G.; Caviness, V.S. Overexpression of p27Kip1, probability of cell cycle exit, and laminar destination of neocortical neurons. Cereb. Cortex 2005, 15, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Nobes, C.D.; Hall, A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef]
- Govek, E.E.; Hatten, M.E.; Van Aelst, L. The role of Rho GTPase proteins in CNS neuronal migration. Dev. Neurobiol. 2011, 71, 528–553. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; He, F.; Kim, K.J.; Blanchi, B.; Coskun, V.; Nguyen, L.; Wu, X.; Zhao, J.; Heng, J.I.; Martinowich, K.; et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. USA 2006, 103, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Ip, J.P.; Ye, T.; Ng, Y.P.; Yung, W.H.; Wu, Z.; Fang, W.; Fu, A.K.; Ip, N.Y. Cdk5-dependent Mst3 phosphorylation and activity regulate neuronal migration through RhoA inhibition. J. Neurosci. 2014, 34, 7425–7436. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liao, G.; Waclaw, R.R.; Burns, K.A.; Linquist, D.; Campbell, K.; Zheng, Y.; Kuan, C.Y. Rac1 controls the formation of midline commissures and the competency of tangential migration in ventral telencephalic neurons. J. Neurosci. 2007, 27, 3884–3893. [Google Scholar] [CrossRef] [PubMed]
- Tahirovic, S.; Hellal, F.; Neukirchen, D.; Hindges, R.; Garvalov, B.K.; Flynn, K.C.; Stradal, T.E.; Chrostek-Grashoff, A.; Brakebusch, C.; Bradke, F. Rac1 regulates neuronal polarization through the WAVE complex. J. Neurosci. 2010, 30, 6930–6943. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, R.; Thumkeo, D.; Kamijo, H.; Kaneko, N.; Sawamoto, K.; Watanabe, K.; Takebayashi, H.; Kiyonari, H.; Ishizaki, T.; Furuyashiki, T.; et al. A role for mDia, a Rho-regulated actin nucleator, in tangential migration of interneuron precursors. Nat. Neurosci. 2012, 15, 373–380, S371–S372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shikanai, M.; Nakajima, K.; Kawauchi, T. N-cadherin regulates radial glial fiber-dependent migration of cortical locomoting neurons. Commun. Integr. Biol. 2011, 4, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Luccardini, C.; Hennekinne, L.; Viou, L.; Yanagida, M.; Murakami, F.; Kessaris, N.; Ma, X.; Adelstein, R.S.; Mege, R.M.; Metin, C. N-cadherin sustains motility and polarity of future cortical interneurons during tangential migration. J. Neurosci. 2013, 33, 18149–18160. [Google Scholar] [CrossRef] [PubMed]
- Luccardini, C.; Leclech, C.; Viou, L.; Rio, J.P.; Metin, C. Cortical interneurons migrating on a pure substrate of N-cadherin exhibit fast synchronous centrosomal and nuclear movements and reduced ciliogenesis. Front. Cell. Neurosci. 2015, 9, 286. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, M.; Miyoshi, R.; Toyokuni, R.; Zhu, Y.; Murakami, F. Dynamics of the leading process, nucleus, and Golgi apparatus of migrating cortical interneurons in living mouse embryos. Proc. Natl. Acad. Sci. USA 2012, 109, 16737–16742. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Wolf, K.; Lammerding, J. Nuclear mechanics during cell migration. Curr. Opin. Cell Biol. 2011, 23, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.; Te Lindert, M.; Krause, M.; Alexander, S.; Te Riet, J.; Willis, A.L.; Hoffman, R.M.; Figdor, C.G.; Weiss, S.J.; Friedl, P. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013, 201, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, K.; Yang, M.; Jiang, P.; Yamamoto, N.; Xu, M.; Amoh, Y.; Tsuji, K.; Bouvet, M.; Tsuchiya, H.; Tomita, K.; et al. Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 2005, 65, 4246–4252. [Google Scholar] [CrossRef] [PubMed]
- Thiam, H.R.; Vargas, P.; Carpi, N.; Crespo, C.L.; Raab, M.; Terriac, E.; King, M.C.; Jacobelli, J.; Alberts, A.S.; Stradal, T.; et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 2016, 7, 10997. [Google Scholar] [CrossRef] [PubMed]
- Jayo, A.; Malboubi, M.; Antoku, S.; Chang, W.; Ortiz-Zapater, E.; Groen, C.; Pfisterer, K.; Tootle, T.; Charras, G.; Gundersen, G.G.; et al. Fascin regulates nuclear movement and deformation in migrating cells. Dev. Cell 2016, 38, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Rivas, R.J.; Hatten, M.E. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J. Neurosci. 1995, 15, 981–989. [Google Scholar] [PubMed]
- Umeshima, H.; Hirano, T.; Kengaku, M. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc. Natl. Acad. Sci. USA 2007, 104, 16182–16187. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Sanada, K.; Samuels, B.A.; Shih, H.; Tsai, L.H. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell 2003, 114, 469–482. [Google Scholar] [CrossRef]
- Itoh, Y.; Higuchi, M.; Oishi, K.; Kishi, Y.; Okazaki, T.; Sakai, H.; Miyata, T.; Nakajima, K.; Gotoh, Y. PDK1-Akt pathway regulates radial neuronal migration and microtubules in the developing mouse neocortex. Proc. Natl. Acad. Sci. USA 2016, 113, E2955–E2964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lei, K.; Yuan, X.; Wu, X.; Zhuang, Y.; Xu, T.; Xu, R.; Han, M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 2009, 64, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Burke, B.; Roux, K.J. Nuclei take a position: Managing nuclear location. Dev. Cell 2009, 17, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Razafsky, D.; Hodzic, D. Bringing KASH under the SUN: The many faces of nucleo-cytoskeletal connections. J. Cell Biol. 2009, 186, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Coffinier, C.; Chang, S.Y.; Nobumori, C.; Tu, Y.; Farber, E.A.; Toth, J.I.; Fong, L.G.; Young, S.G. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 5076–5081. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhang, Z.H.; Yuan, X.B.; Poo, M.M. Spatiotemporal dynamics of traction forces show three contraction centers in migratory neurons. J. Cell Biol. 2015, 209, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Martini, F.J.; Valdeolmillos, M. Actomyosin contraction at the cell rear drives nuclear translocation in migrating cortical interneurons. J. Neurosci. 2010, 30, 8660–8670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.; Zhang, Z.H.; Guan, C.B.; Xia, D.; Yuan, X.B. Leading tip drives soma translocation via forward F-actin flow during neuronal migration. J. Neurosci. 2010, 30, 10885–10898. [Google Scholar] [CrossRef] [PubMed]
- Creppe, C.; Malinouskaya, L.; Volvert, M.L.; Gillard, M.; Close, P.; Malaise, O.; Laguesse, S.; Cornez, I.; Rahmouni, S.; Ormenese, S.; et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 2009, 136, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Laguesse, S.; Creppe, C.; Nedialkova, D.D.; Prevot, P.P.; Borgs, L.; Huysseune, S.; Franco, B.; Duysens, G.; Krusy, N.; Lee, G.; et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell 2015, 35, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Tielens, S.; Huysseune, S.; Godin, J.D.; Chariot, A.; Malgrange, B.; Nguyen, L. Elongator controls cortical interneuron migration by regulating actomyosin dynamics. Cell Res. 2016, 26, 1131–1148. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishimura, Y.V.; Nabeshima, Y.-i.; Kawauchi, T. Morphological and Molecular Basis of Cytoplasmic Dilation and Swelling in Cortical Migrating Neurons. Brain Sci. 2017, 7, 87. https://doi.org/10.3390/brainsci7070087
Nishimura YV, Nabeshima Y-i, Kawauchi T. Morphological and Molecular Basis of Cytoplasmic Dilation and Swelling in Cortical Migrating Neurons. Brain Sciences. 2017; 7(7):87. https://doi.org/10.3390/brainsci7070087
Chicago/Turabian StyleNishimura, Yoshiaki V., Yo-ichi Nabeshima, and Takeshi Kawauchi. 2017. "Morphological and Molecular Basis of Cytoplasmic Dilation and Swelling in Cortical Migrating Neurons" Brain Sciences 7, no. 7: 87. https://doi.org/10.3390/brainsci7070087




