Identification of New Players in Hepatocarcinogenesis: Limits and Opportunities of Using Tissue Microarray (TMA)
Abstract
:1. Introduction: HCC an Overview
2. TMA Methodology: A Historical Overview and Basic Concepts
3. TMA in Hepatocarcinogenesis Research
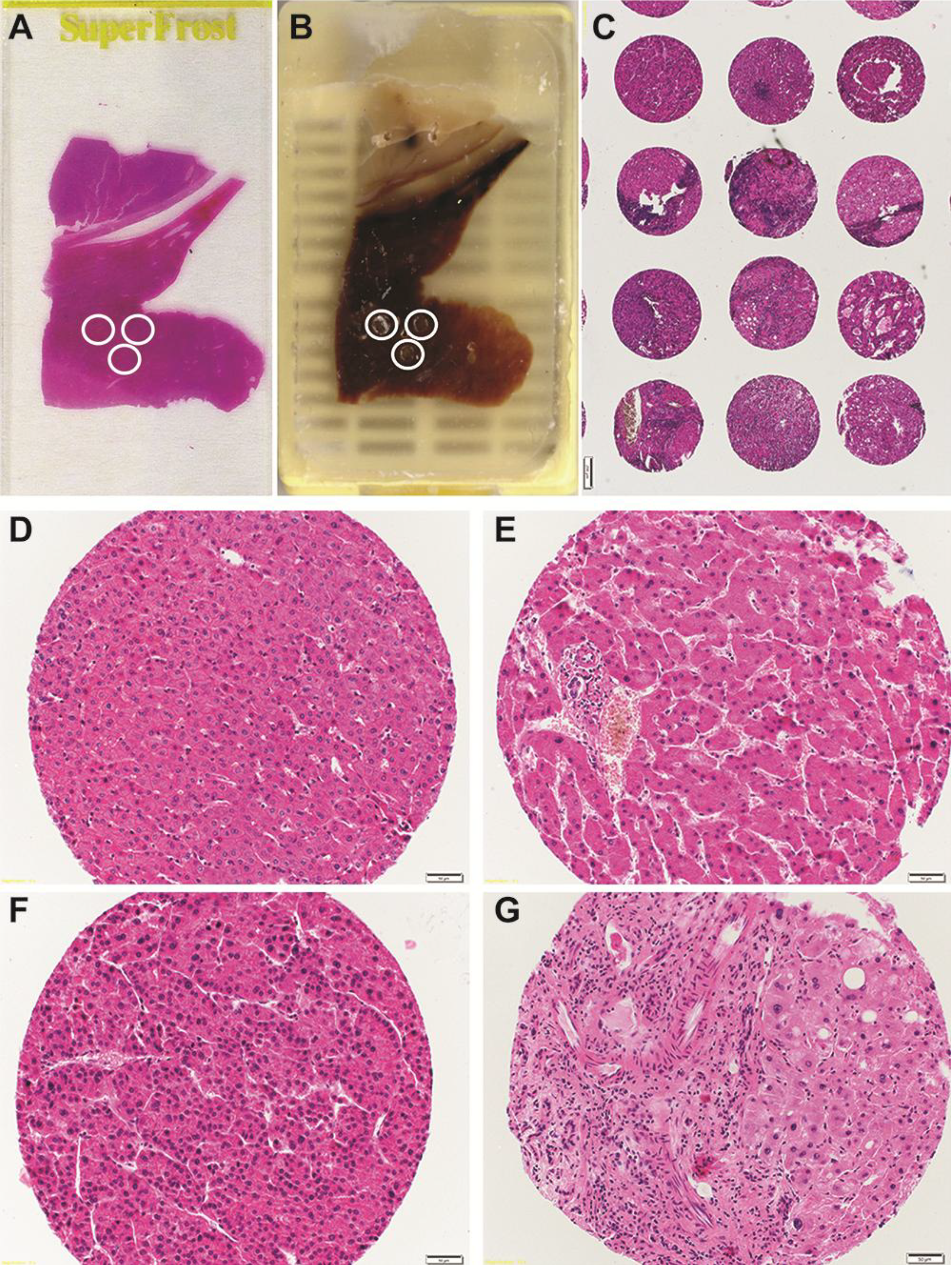
4. Future Prospective
5. Conclusions
Acknowledgments
Author Contributions
Abbreviation
Conflicts of Interest
References
- Gomaa, A.I.; Khan, S.A.; Toledano, M.B.; Waked, I.; Taylor-Robinson, S.D. Hepatocellular carcinoma: Epidemiology, risk factors and pathogenesis. World J. Gastroenterol. 2008, 14, 4300–4308. [Google Scholar] [CrossRef]
- Schutte, K.; Bornschein, J.; Malfertheiner, P. Hepatocellular carcinoma—Epidemiological trends and risk factors. Dig. Dis. 2009, 27, 80–92. [Google Scholar]
- D'Angelo, S.; Secondulfo, M.; de Cristofano, R.; Sorrentino, P. Selection and management of hepatocellular carcinoma patients with sorafenib: Recommendations and opinions from an italian liver unit. Fut. Oncol. 2013, 9, 485–491. [Google Scholar] [CrossRef]
- Marquardt, J.U.; Galle, P.R.; Teufel, A. Molecular diagnosis and therapy of hepatocellular carcinoma (hcc): An emerging field for advanced technologies. J. Hepatol. 2012, 56, 267–275. [Google Scholar] [CrossRef]
- Teufel, A.; Marquardt, J.U.; Staib, F.; Galle, P.R. Snapshot liver transcriptome in hepatocellular carcinoma. J. Hepatol. 2012, 56, 990–992. [Google Scholar] [CrossRef]
- Forner, A.; Llovet, J.M.; Bruix, J. Hepatocellular carcinoma. Lancet 2012, 379, 1245–1255. [Google Scholar] [CrossRef]
- Cucchetti, A.; Piscaglia, F.; Cescon, M.; Ercolani, G.; Pinna, A.D. Systematic review of surgical resection vs. radiofrequency ablation for hepatocellular carcinoma. World J. Gastroenterol. 2013, 19, 4106–4118. [Google Scholar] [CrossRef]
- Welker, M.W.; Bechstein, W.O.; Zeuzem, S.; Trojan, J. Recurrent hepatocellular carcinoma after liver transplantation—An emerging clinical challenge. Transpl. Int. 2013, 26, 109–118. [Google Scholar] [CrossRef]
- Cescon, M.; Cucchetti, A.; Ravaioli, M.; Pinna, A.D. Hepatocellular carcinoma locoregional therapies for patients in the waiting list. Impact on transplantability and recurrence rate. J. Hepatol. 2013, 58, 609–618. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. New Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef]
- Roessler, S.; Long, E.L.; Budhu, A.; Chen, Y.; Zhao, X.; Ji, J.; Walker, R.; Jia, H.L.; Ye, Q.H.; Qin, L.X.; et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 2012, 142, 957–966, e912. [Google Scholar]
- Tornillo, L.; Carafa, V.; Richter, J.; Sauter, G.; Moch, H.; Minola, E.; Gambacorta, M.; Bianchi, L.; Vecchione, R.; Terracciano, L.M. Marked genetic similarities between hepatitis b virus-positive and hepatitis c virus-positive hepatocellular carcinomas. J. Pathol. 2000, 192, 307–312. [Google Scholar] [CrossRef]
- Farazi, P.A.; DePinho, R.A. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat. Rev. Canc. 2006, 6, 674–687. [Google Scholar] [CrossRef]
- Kwon, S.M.; Kim, D.S.; Won, N.H.; Park, S.J.; Chwae, Y.J.; Kang, H.C.; Lee, S.H.; Baik, E.J.; Thorgeirsson, S.S.; Woo, H.G. Genomic copy number alterations with transcriptional deregulation at 6p identify an aggressive hcc phenotype. Carcinogenesis 2013, 34, 1543–1550. [Google Scholar] [CrossRef]
- Oishi, N.; Kumar, M.R.; Roessler, S.; Ji, J.; Forgues, M.; Budhu, A.; Zhao, X.; Andersen, J.B.; Ye, Q.H.; Jia, H.L.; et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of mir-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology 2012, 56, 1792–1803. [Google Scholar] [CrossRef]
- Yang, J.D.; Seol, S.Y.; Leem, S.H.; Kim, Y.H.; Sun, Z.; Lee, J.S.; Thorgeirsson, S.S.; Chu, I.S.; Roberts, L.R.; Kang, K.J. Genes associated with recurrence of hepatocellular carcinoma: Integrated analysis by gene expression and methylation profiling. J. Kor. Med. Sci. 2011, 26, 1428–1438. [Google Scholar] [CrossRef]
- Lee, J.S.; Thorgeirsson, S.S. Comparative and integrative functional genomics of hcc. Oncogene 2006, 25, 3801–3809. [Google Scholar] [CrossRef]
- Wang, X.W.; Thorgeirsson, S.S. Transcriptome analysis of liver cancer: Ready for the clinic? J. Hepatol. 2009, 50, 1062–1064. [Google Scholar] [CrossRef]
- Woo, H.G.; Park, E.S.; Thorgeirsson, S.S.; Kim, Y.J. Exploring genomic profiles of hepatocellular carcinoma. Mol. Carcinog. 2011, 50, 235–243. [Google Scholar] [CrossRef]
- Hoshida, Y.; Toffanin, S.; Lachenmayer, A.; Villanueva, A.; Minguez, B.; Llovet, J.M. Molecular classification and novel targets in hepatocellular carcinoma: Recent advancements. Semin. Liver Dis. 2010, 30, 35–51. [Google Scholar] [CrossRef]
- Battifora, H. The multitumor (sausage) tissue block: Novel method for immunohistochemical antibody testing. Lab. Investig. 1986, 55, 244–248. [Google Scholar]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Barlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998, 4, 844–847. [Google Scholar] [CrossRef]
- Bubendorf, L.; Nocito, A.; Moch, H.; Sauter, G. Tissue microarray (TMA) technology: Miniaturized pathology archives for high-throughput in situ studies. J. Pathol. 2001, 195, 72–79. [Google Scholar] [CrossRef]
- Shergill, I.S.; Shergill, N.K.; Arya, M.; Patel, H.R. Tissue microarrays: A current medical research tool. Curr. Med. Res. Opin. 2004, 20, 707–712. [Google Scholar] [CrossRef]
- Eguiluz, C.; Viguera, E.; Millan, L.; Perez, J. Multitissue array review: A chronological description of tissue array techniques, applications and procedures. Pathol. Res. Pract. 2006, 202, 561–568. [Google Scholar] [CrossRef]
- Chen, W.; Foran, D.J. Advances in cancer tissue microarray technology: Towards improved understanding and diagnostics. Anal. Chim. Acta 2006, 564, 74–81. [Google Scholar] [CrossRef]
- Kallioniemi, O.P.; Kononen, J.; Sauter, G. Introducing tissue microarrays to molecular pathology. Clin. Chem. 2012, 58, 1717–1718. [Google Scholar] [CrossRef]
- Diaz, L.K.; Gupta, R.; Kidwai, N.; Sneige, N.; Wiley, E.L. The use of tma for interlaboratory validation of fish testing for detection of her2 gene amplification in breast cancer. J. Histochem. Cytochem. 2004, 52, 501–507. [Google Scholar] [CrossRef]
- Tzankov, A.; Went, P.; Zimpfer, A.; Dirnhofer, S. Tissue microarray technology: Principles, pitfalls and perspectives—Lessons learned from hematological malignancies. Exp. Gerontol. 2005, 40, 737–744. [Google Scholar] [CrossRef]
- Packeisen, J.; Korsching, E.; Herbst, H.; Boecker, W.; Buerger, H. Demystified...Tissue microarray technology. Mol. Pathol. 2003, 56, 198–204. [Google Scholar] [CrossRef]
- Simon, R.; Nocito, A.; Hubscher, T.; Bucher, C.; Torhorst, J.; Schraml, P.; Bubendorf, L.; Mihatsch, M.M.; Moch, H.; Wilber, K.; et al. Patterns of her-2/neu amplification and overexpression in primary and metastatic breast cancer. J. Natl. Canc. Inst. 2001, 93, 1141–1146. [Google Scholar] [CrossRef]
- Schraml, P.; Kononen, J.; Bubendorf, L.; Moch, H.; Bissig, H.; Nocito, A.; Mihatsch, M.J.; Kallioniemi, O.P.; Sauter, G. Tissue microarrays for gene amplification surveys in many different tumor types. Clin. Canc. Res. 1999, 5, 1966–1975. [Google Scholar]
- Hoos, A.; Urist, M.J.; Stojadinovic, A.; Mastorides, S.; Dudas, M.E.; Leung, D.H.; Kuo, D.; Brennan, M.F.; Lewis, J.J.; Cordon-Cardo, C. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am. J. Pathol. 2001, 158, 1245–1251. [Google Scholar] [CrossRef]
- Skacel, M.; Skilton, B.; Pettay, J.D.; Tubbs, R.R. Tissue microarrays: A powerful tool for high-throughput analysis of clinical specimens: A review of the method with validation data. Appl. Immunohistochem. Mol. Morphol. 2002, 10, 1–6. [Google Scholar] [CrossRef]
- Lau, S.H.; Sham, J.S.; Xie, D.; Tzang, C.H.; Tang, D.; Ma, N.; Hu, L.; Wang, Y.; Wen, J.M.; Xiao, G.; et al. Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene 2006, 25, 1242–1250. [Google Scholar] [CrossRef]
- Kazakova, M.H.; Sarafian, V.S. Ykl-40—A novel biomarker in clinical practice? Folia Med. 2009, 51, 5–14. [Google Scholar]
- Hu, S.; Zhang, M.; Lv, Z.; Bi, J.; Dong, Y.; Wen, J. Expression of zinc-fingers and homeoboxes 2 in hepatocellular carcinogenesis: A tissue microarray and clinicopathological analysis. Neoplasma 2007, 54, 207–211. [Google Scholar]
- Hu, L.; Sham, J.S.; Xie, D.; Wen, J.M.; Wang, W.S.; Wang, Y.; Guan, X.Y. Up-regulation of fibroblast growth factor 3 is associated with tumor metastasis and recurrence in human hepatocellular carcinoma. Canc. Lett. 2007, 252, 36–42. [Google Scholar] [CrossRef]
- Chen, G.; Dang, Y.W.; Luo, D.Z.; Feng, Z.B.; Tang, X.L. Expression of heparanase in hepatocellular carcinoma has prognostic significance: A tissue microarray study. Oncol. Res. 2008, 17, 183–189. [Google Scholar] [CrossRef]
- Shengbing, Z.; Feng, L.J.; Bin, W.; Lingyun, G.; Aimin, H. Expression of kiss-1 gene and its role in invasion and metastasis of human hepatocellular carcinoma. Anat. Rec. 2009, 292, 1128–1134. [Google Scholar] [CrossRef]
- Yang, X.R.; Xu, Y.; Yu, B.; Zhou, J.; Qiu, S.J.; Shi, G.M.; Zhang, B.H.; Wu, W.Z.; Shi, Y.H.; Wu, B.; et al. High expression levels of putative hepatic stem/progenitor cell biomarkers related to tumour angiogenesis and poor prognosis of hepatocellular carcinoma. Gut 2010, 59, 953–962. [Google Scholar] [CrossRef]
- Bae, J.S.; Choi, H.N.; Noh, S.J.; Park, B.H.; Jang, K.Y.; Park, C.K.; Moon, W.S. Expression of k19 and k7 in dysplastic nodules and hepatocellular carcinoma. Oncol. Lett. 2012, 4, 213–220. [Google Scholar]
- Cheng, A.S.; Lau, S.S.; Chen, Y.; Kondo, Y.; Li, M.S.; Feng, H.; Ching, A.K.; Cheung, K.F.; Wong, H.K.; Tong, J.H.; et al. Ezh2-mediated concordant repression of wnt antagonists promotes beta-catenin-dependent hepatocarcinogenesis. Canc. Res. 2011, 71, 4028–4039. [Google Scholar] [CrossRef]
- Geraud, C.; Mogler, C.; Runge, A.; Evdokimov, K.; Lu, S.; Schledzewski, K.; Arnold, B.; Hammerling, G.; Koch, P.S.; Breuhahn, K.; et al. Endothelial transdifferentiation in hepatocellular carcinoma: Loss of stabilin-2 expression in peri-tumourous liver correlates with increased survival. Liver Int. 2013, 33, 1428–1440. [Google Scholar] [CrossRef]
- Baumhoer, D.; Tornillo, L.; Stadlmann, S.; Roncalli, M.; Diamantis, E.K.; Terracciano, L.M. Glypican 3 expression in human nonneoplastic, preneoplastic, and neoplastic tissues: A tissue microarray analysis of 4,387 tissue samples. Am. J. Clin. Pathol. 2008, 129, 899–906. [Google Scholar] [CrossRef]
- Fasano, M.; Theise, N.D.; Nalesnik, M.; Goswami, S.; Garcia de Davila, M.T.; Finegold, M.J.; Greco, M.A. Immunohistochemical evaluation of hepatoblastomas with use of the hepatocyte-specific marker, hepatocyte paraffin 1, and the polyclonal anti-carcinoembryonic antigen. Mod. Pathol. 1998, 11, 934–938. [Google Scholar]
- Lugli, A.; Tornillo, L.; Mirlacher, M.; Bundi, M.; Sauter, G.; Terracciano, L.M. Hepatocyte paraffin 1 expression in human normal and neoplastic tissues: Tissue microarray analysis on 3,940 tissue samples. Am. J. Clin. Pathol. 2004, 122, 721–727. [Google Scholar] [CrossRef]
- Song, T.J.; Fong, Y.; Cho, S.J.; Gonen, M.; Hezel, M.; Tuorto, S.; Choi, S.Y.; Kim, Y.C.; Suh, S.O.; Koo, B.H.; et al. Comparison of hepatocellular carcinoma in american and asian patients by tissue array analysis. J. Surg. Oncol. 2012, 106, 84–88. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Solomon, M.; Satyamoorthy, K.; Martin, L.E.; Lingen, M.W. Tissue microarray—A high-throughput molecular analysis in head and neck cancer. J. Oral Pathol. Med. 2008, 37, 166–176. [Google Scholar]
- Torhorst, J.; Bucher, C.; Kononen, J.; Haas, P.; Zuber, M.; Kochli, O.R.; Mross, F.; Dieterich, H.; Moch, H.; Mihatsch, M.; et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am. J. Pathol. 2001, 159, 2249–2256. [Google Scholar] [CrossRef]
- Bubendorf, L.; Kononen, J.; Koivisto, P.; Schraml, P.; Moch, H.; Gasser, T.C.; Willi, N.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Canc. Res. 1999, 59, 803–806. [Google Scholar]
- Bubendorf, L.; Kolmer, M.; Kononen, J.; Koivisto, P.; Mousses, S.; Chen, Y.; Mahlamaki, E.; Schraml, P.; Moch, H.; Willi, N.; et al. Hormone therapy failure in human prostate cancer: Analysis by complementary DNA and tissue microarrays. J. Natl. Canc. Inst. 1999, 91, 1758–1764. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Quagliata, L.; Schlageter, M.; Quintavalle, C.; Tornillo, L.; Terracciano, L.M. Identification of New Players in Hepatocarcinogenesis: Limits and Opportunities of Using Tissue Microarray (TMA). Microarrays 2014, 3, 91-102. https://doi.org/10.3390/microarrays3020091
Quagliata L, Schlageter M, Quintavalle C, Tornillo L, Terracciano LM. Identification of New Players in Hepatocarcinogenesis: Limits and Opportunities of Using Tissue Microarray (TMA). Microarrays. 2014; 3(2):91-102. https://doi.org/10.3390/microarrays3020091
Chicago/Turabian StyleQuagliata, Luca, Manuel Schlageter, Cristina Quintavalle, Luigi Tornillo, and Luigi M. Terracciano. 2014. "Identification of New Players in Hepatocarcinogenesis: Limits and Opportunities of Using Tissue Microarray (TMA)" Microarrays 3, no. 2: 91-102. https://doi.org/10.3390/microarrays3020091



