Gut Microbiota-Dependent Trimethylamine-N-oxide and Serum Biomarkers in Patients with T2DM and Advanced CKD
Abstract
:1. Introduction
2. Materials and Methods
2.1. Research Subjects
2.2. DNA Extraction from Stools and Sequencing
2.3. TMAO Assay
2.4. Biomarker Assays
2.5. Statistical Methods
3. Results
3.1. Anthropometric and Metabolic Parameters in the Study Groups
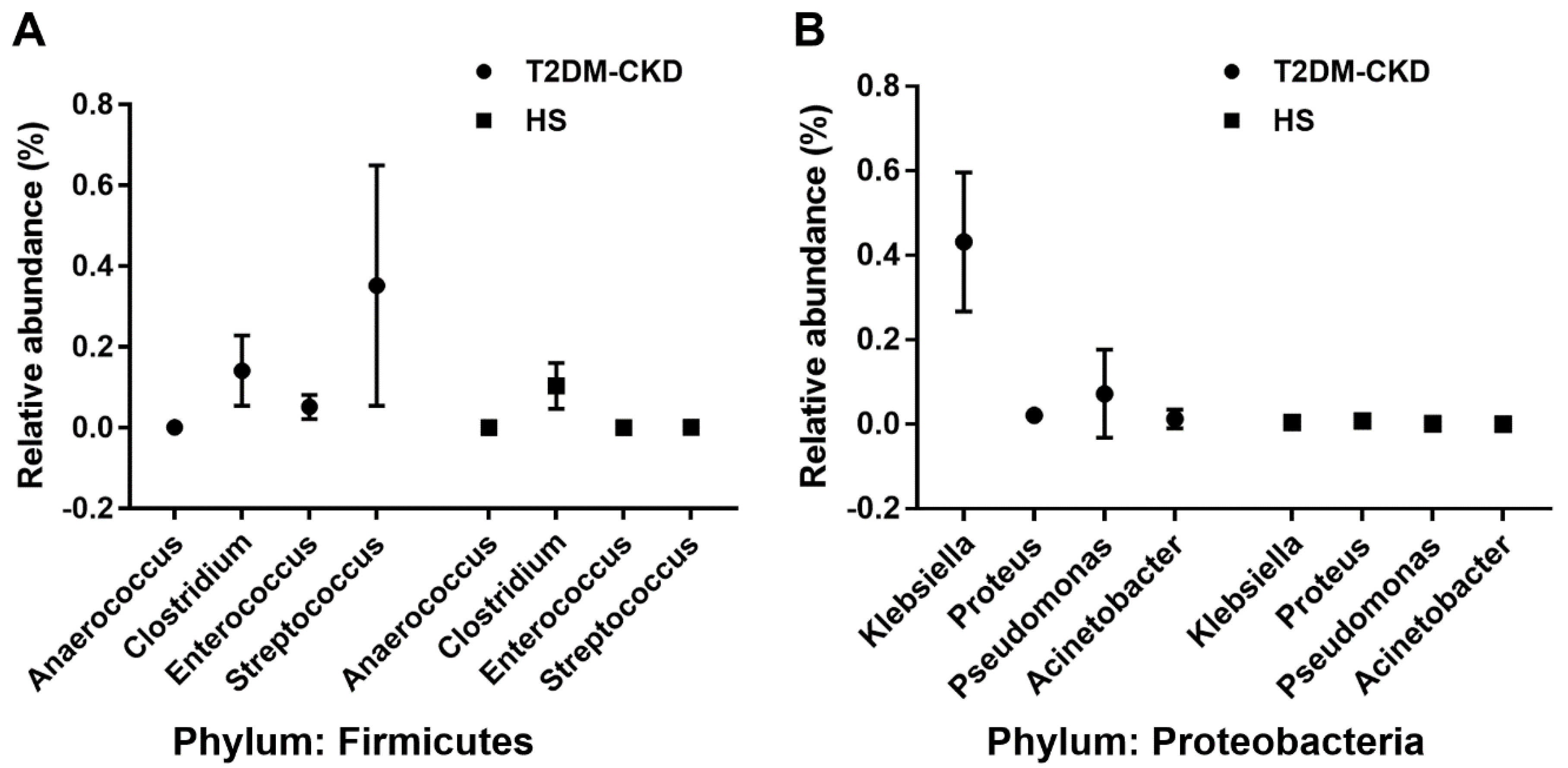
3.2. TMA Gut Producing Bacteria in T2DM-CKD Patients
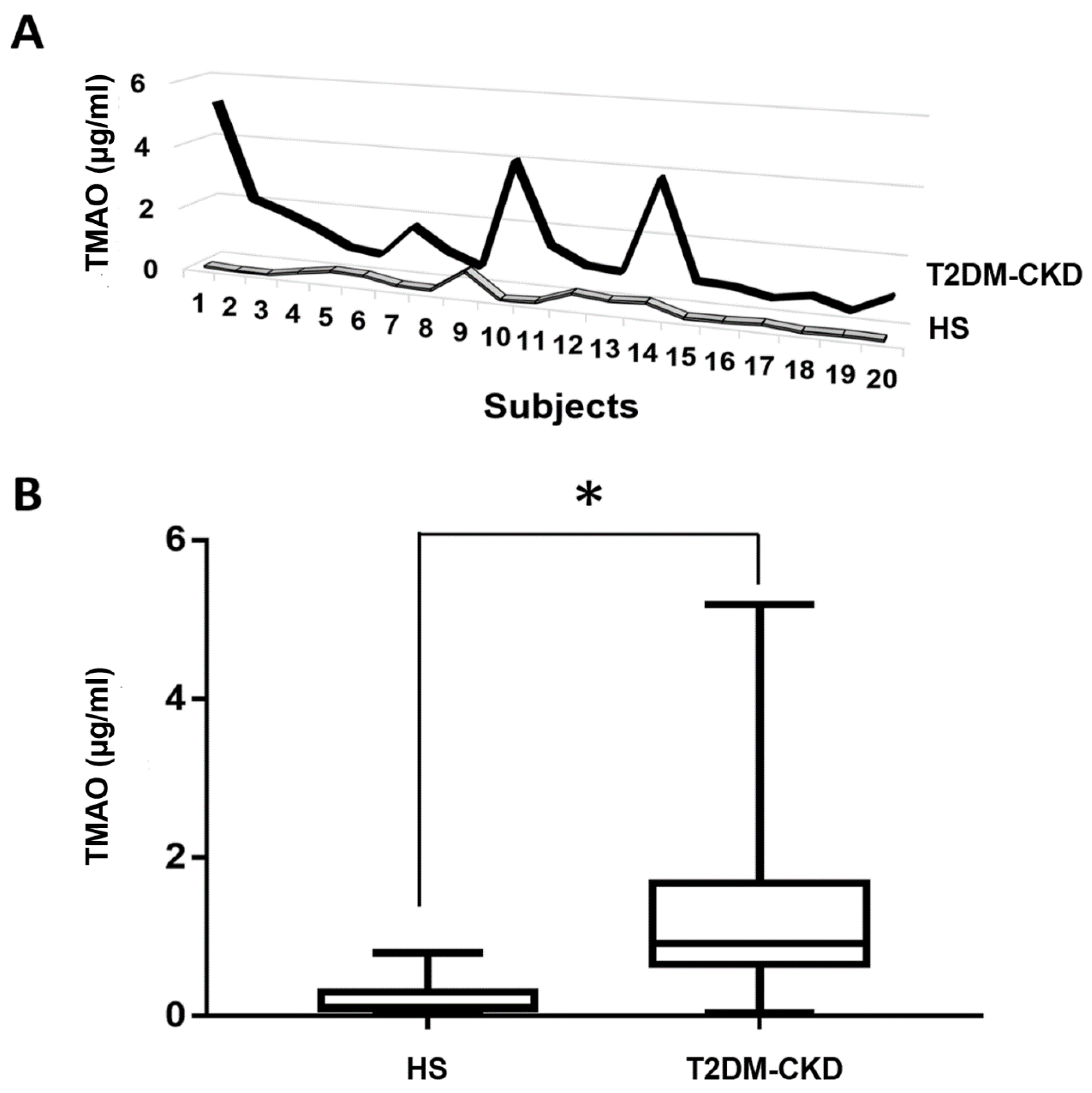
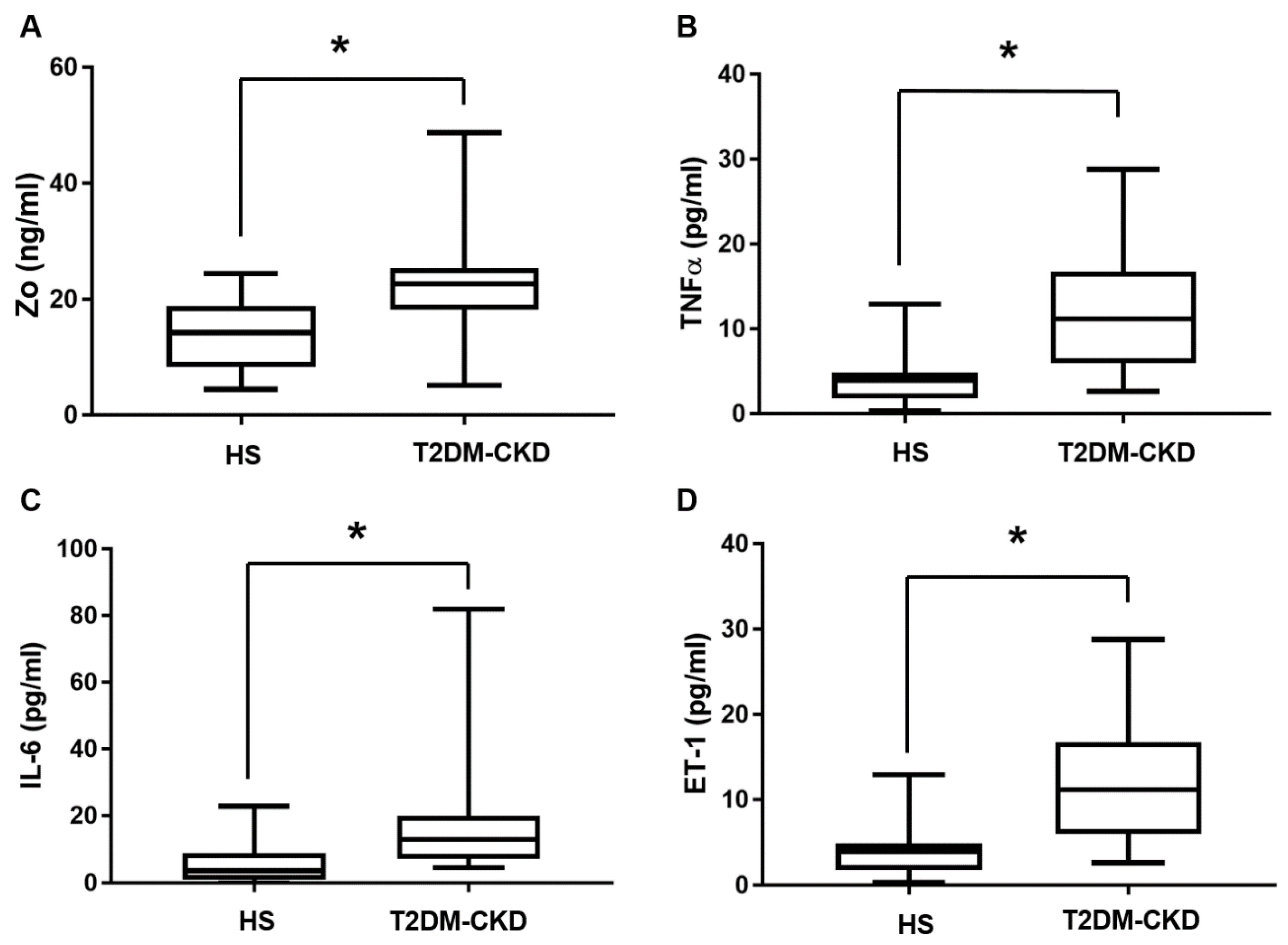
3.3. TMAO and Serum Biomarkers of Inflammation and Endothelial Dysfunction Levels in T2DM-CKD Patients and Healthy Subjects
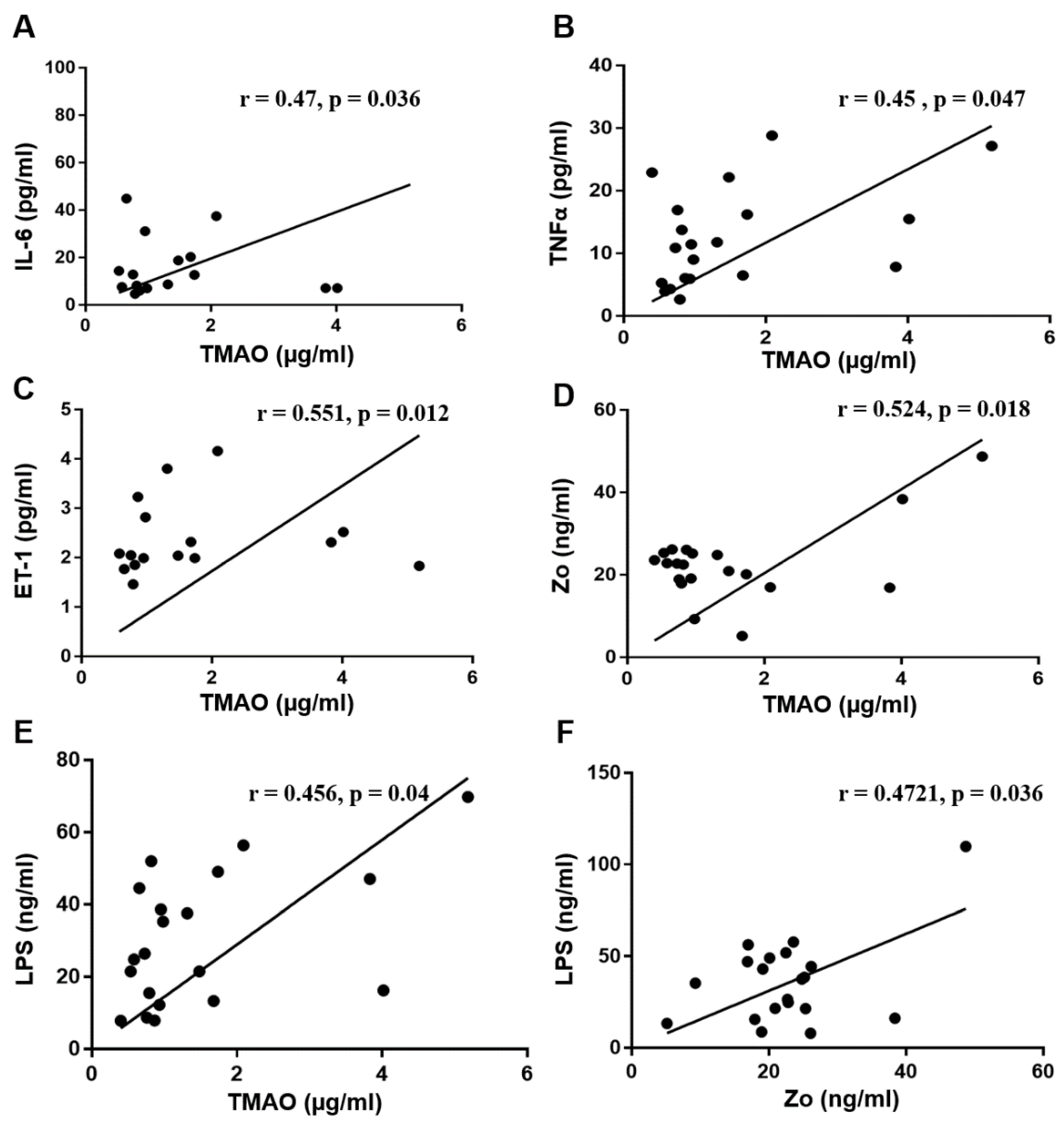
3.4. Correlation Analysis between Serum TMAO and Serum Biomarkers
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hill, N.R.; Fatoba, S.T.; Oke, J.L.; Hirst, J.A.; O’Callaghan, C.A.; Lasserson, D.S.; Hobbs, F.D. Global Prevalence of Chronic Kidney Disease—A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0158765. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.C.; Cooper, M.E.; Zimmet, P. Changing epidemiology of type 2 diabetes mellitus and associated chronic kidney disease. Nat. Rev. Nephrol. 2016, 12, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, C.; Hu, C.; Han, Y.; Zhao, L.; Zhu, X.; Xiao, L.; Sun, L. Normoalbuminuric diabetic kidney disease. Front. Med. 2017, 11, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Olsen, S. Identification of non-diabetic glomerular disease in renal biopsies from diabetics—A dilemma. Nephrol. Dial. Transplant. 1999, 14, 1846–18499. [Google Scholar] [CrossRef] [PubMed]
- Pálsson, R.; Patel, U.D. Cardiovascular complications of diabetic kidney disease. Adv. Chronic Kidney Dis. 2014, 21, 273–280. [Google Scholar] [CrossRef]
- Shikata, K.; Makino, H. Microinflammation in the pathogenesis of diabetic nephropathy. J. Diabetes Investig. 2013, 4, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Duran-Salgado, M.B.; Rubio-Guerra, A.F. Diabetic nephropathy and inflammation. World J. Diabetes 2014, 5, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, A.; Massy, Z.A.; Meijers, B.; Evenepoel, P.; Vanholder, R.; Raj, D.S. Role of the gut microbiome in uremia: A potential therapeutic target. Am. J. Kidney Dis. 2016, 67, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Wing, M.R.; Patel, S.S.; Ramezani, A.; Raj, D.S. Gut microbiome in chronic kidney disease. Exp. Physiol. 2016, 101, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Nallu, A.; Sharma, S.; Ramezani, A.; Muralidharan, J.; Raj, D. Gut microbiome in chronic kidney disease: Challenges and opportunities. Transl. Res. 2017, 179, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.; Xia, Y. Further analysis reveals new gut microbiome markers of type 2 diabetes mellitus. Antonie Van Leeuwenhoek 2017, 110, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability—A new target for disease prevention and therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef] [PubMed]
- Nesseler, N.; Launey, Y.; Aninat, C.; More, L.F.; Mallédant, Y.; Seguin, P. Clinical review: The liver in sepsis. Crit. Care 2012, 16, 235. [Google Scholar] [CrossRef] [PubMed]
- Haak, B.W.; Wiersinga, W.J. The role of the gut microbiota in sepsis. Lancet Gastroenterol. Hepatol. 2017, 2, 135–143. [Google Scholar] [CrossRef]
- Chhibber-Goel, J.; Gaur, A.; Singhal, V.; Parakh, N.; Bhargava, B.; Sharma, A. The complex metabolism of trimethylamine in humans: Endogenous and exogenous sources. Expert Rev. Mol. Med. 2016, 18, e8. [Google Scholar] [CrossRef] [PubMed]
- Phillips, I.R.; Dolphin, C.T.; Clair, P.; Hadley, M.R.; Hutt, A.J.; McCombie, R.R.; Smith, R.L.; Shephard, E.A. The molecular biology of the flavin-containing monooxygenases of man. Chem. Biol. Interact. 1995, 96, 17–32. [Google Scholar] [CrossRef]
- Lawton, M.P.; Cashman, J.R.; Cresteil, T.; Dolphin, C.T.; Elfarra, A.A.; Hines, R.N.; Hodgson, E.; Kimura, T.; Ozols, J.; Phillips, I.R.; et al. Arch. A nomenclature for the mammalian flavin-containing monooxygenase gene family based on amino acid sequence identities. Biochem. Biophys. 1994, 308, 254–257. [Google Scholar] [CrossRef]
- Hernandez, D.; Janmohamed, A.; Chandan, P.; Phillips, I.R.; Shephard, E.A. Organization and evolution of the flavin-containing monooxygenase genes of human and mouse: Identification of novel gene and pseudogene clusters. Pharmacogenetics 2004, 14, 17–130. [Google Scholar] [CrossRef] [PubMed]
- Hines, R.N.; Luo, Z.; Hopp, K.A.; Cabacungan, E.T.; Koukouritaki, S.B.; McCarver, D.G. Genetic variability at the human FMO1 locus: Significance of a basal promoter yin yang 1 element polymorphism (FMO1*6). J. Pharmacol. Exp. Ther. 2003, 306, 1210–1218. [Google Scholar] [CrossRef] [PubMed]
- Cashman, J.R.; Zhang, J. Human flavin-containing monooxygenases. Annu. Rev. Pharmacol. Toxicol. 2006, 46, 65–100. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jiao, X.; Ma, Y.; Liu, Y.; Zhang, L.; He, Y.; Chen, Y. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome. Biochem. Biophys. Res. Commun. 2016, 481, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Schugar, R.C.; Brown, J.M. Emerging roles of flavin monooxygenase 3 in cholesterol metabolism and atherosclerosis. Curr. Opin. Lipidol. 2015, 26, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Ling, A.V.; Manthena, P.V.; Gearing, M.E.; Graham, M.J.; Crooke, R.M.; Croce, K.J.; Esquejo, R.; Clish, C.B.; Morbid Obesity Study Group; et al. Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat. Commun. 2015, 6, 6498. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Davignon, J.; Ganz, P. Role of endothelial dysfunction in atherosclerosis. Circulation 2004, 109, III27–III32. [Google Scholar] [CrossRef] [PubMed]
- Manfrini, O.; Cenko, E.; Verna, E.; Salerno Uriarte, J.A.; Bugiardini, R. Endothelial dysfunction versus early atherosclerosis: A study with high resolution imaging. Int. J. Cardiol. 2013, 168, 1714–1716. [Google Scholar] [CrossRef] [PubMed]
- Warrier, M.; Shih, D.M.; Burrows, A.C.; Ferguson, D.; Gromovsky, A.D.; Brown, A.L.; Marshall, S.; McDaniel, A.; Schugar, R.C.; Wang, Z.; et al. The TMAO-Generating enzyme flavin monooxygenase 3 is a central regulator of cholesterol balance. Cell Rep. 2015, 10, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Seldin, M.M.; Meng, Y.; Qi, H.; Zhu, W.; Wang, Z.; Hazen, S.L.; Lusis, A.J.; Shih, D.M. Trimethylamine N-Oxide promotes vascular inflammation through signaling of mitogen-activated protein kinase and nuclear factor-κB. J. Am. Heart Assoc. 2016, 5, e002767. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Pan, B.; Chen, Y.; Guo, C.; Zhao, M.; Zheng, L.; Chen, B. Trimethylamine N-oxide in atherogenesis: Impairing endothelial self-repair capacity and enhancing monocyte adhesion. Biosci. Rep. 2017, 37, BSR20160244. [Google Scholar] [CrossRef] [PubMed]
- Richesson, R.L.; Rusincovitch, S.A.; Wixted, D.; Batch, B.C.; Feinglos, M.N.; Miranda, M.L.; Hammond, W.E.; Califf, R.M.; Spratt, S.E. A comparison of phenotype definitions for diabetes mellitus. J. Am. Med. Inform. Assoc. 2013, 20, e319–e326. [Google Scholar] [CrossRef] [PubMed]
- Fennema, D.; Phillips, I.R.; Shephard, E.A. Trimethylamine and Trimethylamine N-Oxide, a flavin-containing monooxygenase 3 (fmo3)-mediated host-microbiome metabolic axis implicated in health and disease. Drug Metab. Dispos. 2016, 44, 1839–1850. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Hazen, S.L. The contributory role of gut microbiota in cardiovascular disease. J. Clin. Investig. 2014, 124, 4204–4211. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Miele, L.; Giorgio, V.; Alberelli, M.A.; De Candia, E.; Gasbarrini, A.; Grieco, A. Impact of gut microbiota on obesity, diabetes, and cardiovascular disease risk. Curr. Cardiol. Rep. 2015, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Fu, Y.; Lv, Z. Association of Helicobacter pylori infection with diabetic complications: A meta-analysis. Endocr. Res. 2014, 39, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhu, S.; Xu, G. Targeting gut microbiota: A potential promising therapy for diabetic kidney disease. Am. J. Transl. Res. 2016, 8, 4009–4016. [Google Scholar] [PubMed]
- Ficek, J.; Wyskida, K.; Ficek, R.; Wajda, J.; Klein, D.; Witkowicz, J.; Rotkegel, S.; Spiechowicz-Zatoń, U.; Kocemba-Dyczek, J.; Ciepał, J.; et al. Relationship between plasma levels of zonulin, bacterial lipopolysaccharides, D-lactate and markers of inflammation in haemodialysis patients. Int. Urol. Nephrol. 2017, 49, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Sturgeon, C.; Fasano, A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue Barriers 2016, 4, e1251384. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. Zonulin and its regulation of intestinal barrier function: The biological door to inflammation, autoimmunity, and cancer. Physiol. Rev. 2011, 91, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Wang, Z.; Kennedy, D.J.; Wu, Y.; Buffa, J.A.; Agatisa-Boyle, B.; Li, X.S.; Levison, B.S.; Hazen, S.L. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015, 116, 448–455, PMCID:PMC4312512. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, S.; Linseisen, J.; Allenspach, M.; von Eckardstein, A.; Müller, D. Plasma Concentrations of Trimethylamine-N-oxide are directly associated with dairy food consumption and low-grade inflammation in a german adult population. J. Nutr. 2016, 146, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Jayashree, B.; Bibin, Y.S.; Prabhu, D.; Shanthirani, C.S.; Gokulakrishnan, K.; Lakshmi, B.S.; Mohan, V.; Balasubramanyam, M. Increased circulatory levels of lipopolysaccharide (LPS) and zonulin signify novel biomarkers of proinflammation in patients with type 2 diabetes. Mol. Cell Biochem. 2014, 388, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Mokkala, K.; Röytiö, H.; Munukka, E.; Pietilä, S.; Ekblad, U.; Rönnemaa, T.; Eerola, E.; Laiho, A.; Laitinen, K. Gut Microbiota richness and composition and dietary intake of overweight pregnant women are related to serum zonulin concentration, a marker for intestinal permeability. J. Nutr. 2016, 146, 1694–1700. [Google Scholar] [CrossRef] [PubMed]
- Dschietzig, T.B.; Boschann, F.; Ruppert, J.; Armbruster, F.P.; Meinitzer, A.; Bankovic, D.; Mitrovic, V.; Melzer, C. Plasma zonulin and its association with kidney function, severity of heart failure, and metabolic inflammation. Clin. Lab. 2016, 62, 2443–2447. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Salvatore, F. The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta 2015, 451, 97–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.Q.; Mitchell, S.C.; Smith, R.L. Dietary precursors of trimethylamine in man: A pilot study. Food Chem. Toxicol. 1999, 37, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Ussher, J.R.; Lopaschuk, G.D.; Arduini, A. Gut microbiota metabolism of L-carnitine and cardiovascular risk. Atherosclerosis 2013, 231, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Boutagy, N.E.; Neilson, A.P.; Osterberg, K.L.; Smithson, A.T.; Englund, T.R.; Davy, B.M.; Hulver, M.W.; Davy, K.P. Short-term high-fat diet increases postprandial trimethylamine-N-oxide in humans. Nutr. Res. 2015, 35, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Falcinelli, S.; Rodiles, A.; Hatef, A.; Picchietti, S.; Cossignani, L.; Merrifield, D.L.; Unniappan, S.; Carnevali, O. Dietary lipid content reorganizes gut microbiota and probiotic L. rhamnosus attenuates obesity and enhances catabolic hormonal milieu in zebrafish. Sci. Rep. 2017, 7, 5512. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Sonne, S.B.; Feng, Q.; Chen, N.; Xia, Z.; Li, X.; Fang, Z.; Zhang, D.; Fjære, E.; Midtbø, L.K.; et al. High-fat feeding rather than obesity drives taxonomical and functional changes in the gut microbiota in mice. Microbiome 2017, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Stone, N.J.; Ballantyne, C.; Bittner, V.; Criqui, M.H.; Ginsberg, H.N.; Goldberg, A.C.; Howard, W.J.; Jacobson, M.S.; Kris-Etherton, P.M.; et al. Triglycerides and cardiovascular disease: A scientific statement from the american heart association. Circulation 2011, 123, 2292–2333. [Google Scholar] [CrossRef] [PubMed]
- Nordestgaard, B.G. Triglyceride-rich lipoproteins and atherosclerotic cardiovascular disease: New insights from epidemiology, genetics, and biology. Circ. Res. 2016, 118, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Obeid, R.; Awwad, H.M.; Rabagny, Y.; Graeber, S.; Herrmann, W.; Geisel, J. Plasma trimethylamine N-oxide concentration is associated with choline, phospholipids, and methyl metabolism. Am. J. Clin. Nutr. 2016, 103, 703–711. [Google Scholar] [CrossRef] [PubMed]

| Parameters | HS | T2DM-CKD | p-Value |
|---|---|---|---|
| Age (Years) | 54.3 ± 3.2 | 64.4 ± 2.3 | NS |
| BMI (kg/m2) | 28.17 ± 1.1 | 33.2 ± 2.9 | NS |
| Total cholesterol (mg/dL) | 195 ± 11 | 175.7 ± 13 | <0.05 |
| LDL (mg/dL) | 72 ± 3.2 | 99 ± 10 | <0.03 |
| HDL (mg/dL) | 58 ± 3.3 | 37.7 ± 3.0 | <0.05 |
| Triglyceride (mg/dL) | 85 ± 4.1 | 206.9 ± 21 | <0.001 |
| Hemoglobin (g/dL) | 11.9 ± 0.2 | 12.3 ± 0.4 | NS |
| * Protein (%) | 17 | 12 | <0.05 |
| * Fat (%) | 32.7 | 50.0 | <0.03 |
| * Carbohydrate (%) | 52 | 47 | NS |
| Genus | T2DM-CKD (%) | HS (%) |
|---|---|---|
| Bifidobacterium ꜜ,* | 0.459 | 0.565 |
| Clostridium ꜛ,* | 0.114 | 0.102 |
| Escherichia ꜛ,* | 0.0007 | 0.0001 |
| Enterobacter ꜛ,* | 0.0004 | <10−5 |
| Acinetobacter ꜛ,* | 0.0105 | <10−5 |
| Proteus ꜛ,* | 0.0142 | <10−5 |
| Lactobacillus ꜛ,* | 1.03 | 0.046 |
| Phylum (Out of 20 Total) | Family (Out of 158 total) | Genus (Out of 357 Total) | T2DM-CKD (%) | HS (%) |
|---|---|---|---|---|
| Firmicutes | Tissierellaceae | Anaerococcus *,ꜛ | 0.001 | 0.00003 |
| Clostridiaceae | Clostridium *,ꜛ | 0.114 | 0.102 | |
| Peptococcaceae | Desulfitobacter *,ꜛ | 0.00023 | <10−5 | |
| Enterococcaceae | Enterococcus *,ꜛ | 0.052 | 0.00025 | |
| Streptococcaceae | Streptococcus *,ꜛ | 0.368 | 0.0013 | |
| Proteobacteria | Desulfovibrionaceae | Desulfovibrio *,ꜛ | 0.186 | 0.111 |
| Enterobacteriaceae | Enterobacter *,ꜛ | 0.0004 | <10−5 | |
| Enterobacteriaceae | Escherichia *,†,ꜛ | 0.0007 | 0.0001 | |
| Enterobacteriaceae | Klebsiella *,†,ꜛ | 0.433 | 0.0086 | |
| Enterobacteriaceae | Proteus *,†,ꜛ | 0.0142 | <10−5 | |
| Pseudomonadaceae | Pseudomonas *,†,ꜛ | 0.021 | 0.0012 | |
| Moraxellaceae | Acinetobacter †,ꜛ | 0.0105 | <10−5 | |
| Enterobacteriaceae | Citrobacter †,ꜛ | 0.00088 | 0.00007 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Obaide, M.A.I.; Singh, R.; Datta, P.; Rewers-Felkins, K.A.; Salguero, M.V.; Al-Obaidi, I.; Kottapalli, K.R.; Vasylyeva, T.L. Gut Microbiota-Dependent Trimethylamine-N-oxide and Serum Biomarkers in Patients with T2DM and Advanced CKD. J. Clin. Med. 2017, 6, 86. https://doi.org/10.3390/jcm6090086
Al-Obaide MAI, Singh R, Datta P, Rewers-Felkins KA, Salguero MV, Al-Obaidi I, Kottapalli KR, Vasylyeva TL. Gut Microbiota-Dependent Trimethylamine-N-oxide and Serum Biomarkers in Patients with T2DM and Advanced CKD. Journal of Clinical Medicine. 2017; 6(9):86. https://doi.org/10.3390/jcm6090086
Chicago/Turabian StyleAl-Obaide, Mohammed A. I., Ruchi Singh, Palika Datta, Kathy A. Rewers-Felkins, Maria V. Salguero, Ibtisam Al-Obaidi, Kameswara Rao Kottapalli, and Tetyana L. Vasylyeva. 2017. "Gut Microbiota-Dependent Trimethylamine-N-oxide and Serum Biomarkers in Patients with T2DM and Advanced CKD" Journal of Clinical Medicine 6, no. 9: 86. https://doi.org/10.3390/jcm6090086





