Near Infrared Spectrometry for Rapid Non-Invasive Modelling of Aspergillus-Contaminated Maturing Kernels of Maize (Zea mays L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Maize Crop Development and Preparation of Kernels and Fungal Spore Suspensions
2.2. Near Infrared Spectral Data from Maize Kernels and Treatment of Spectral Data
3. Results
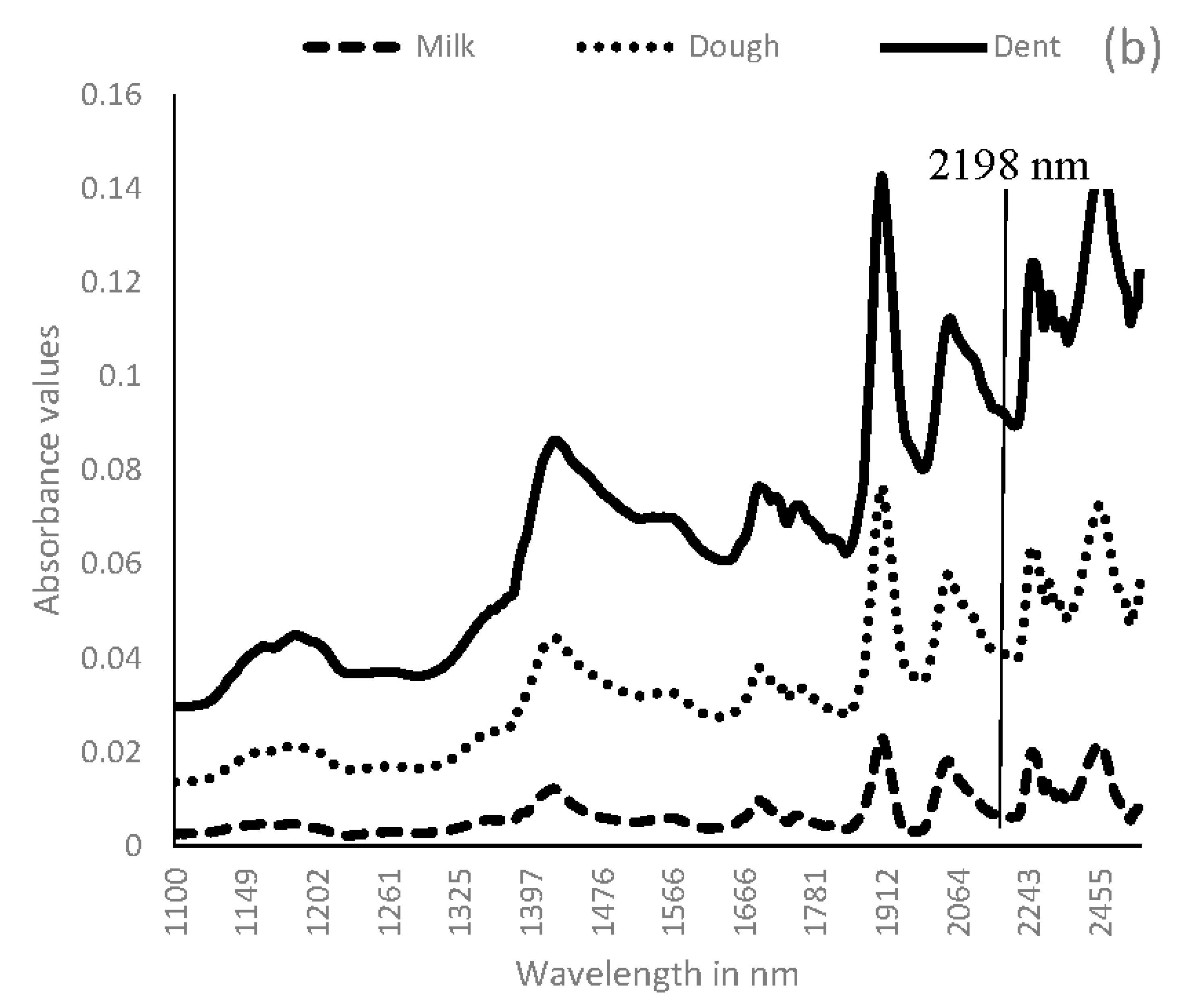
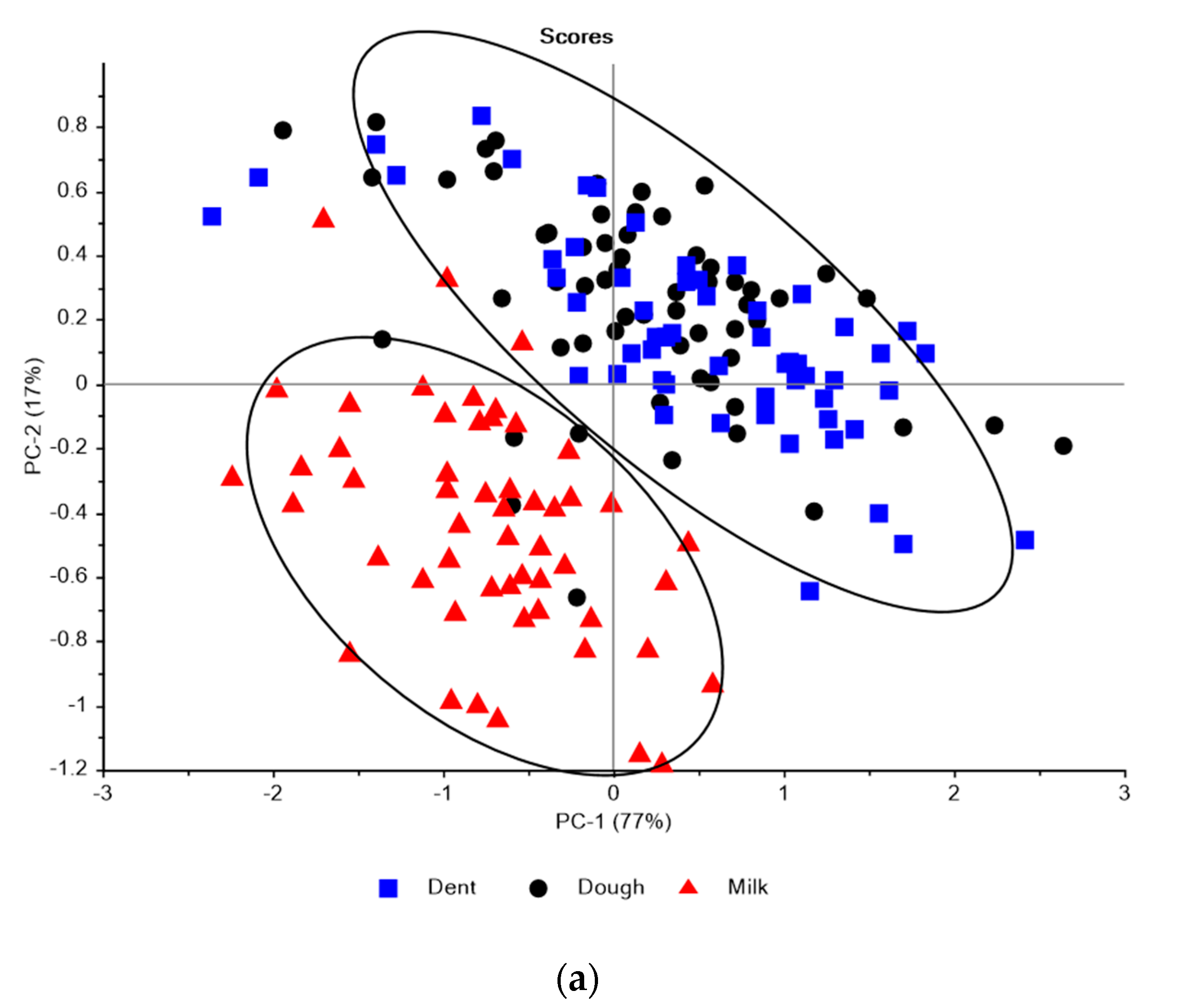
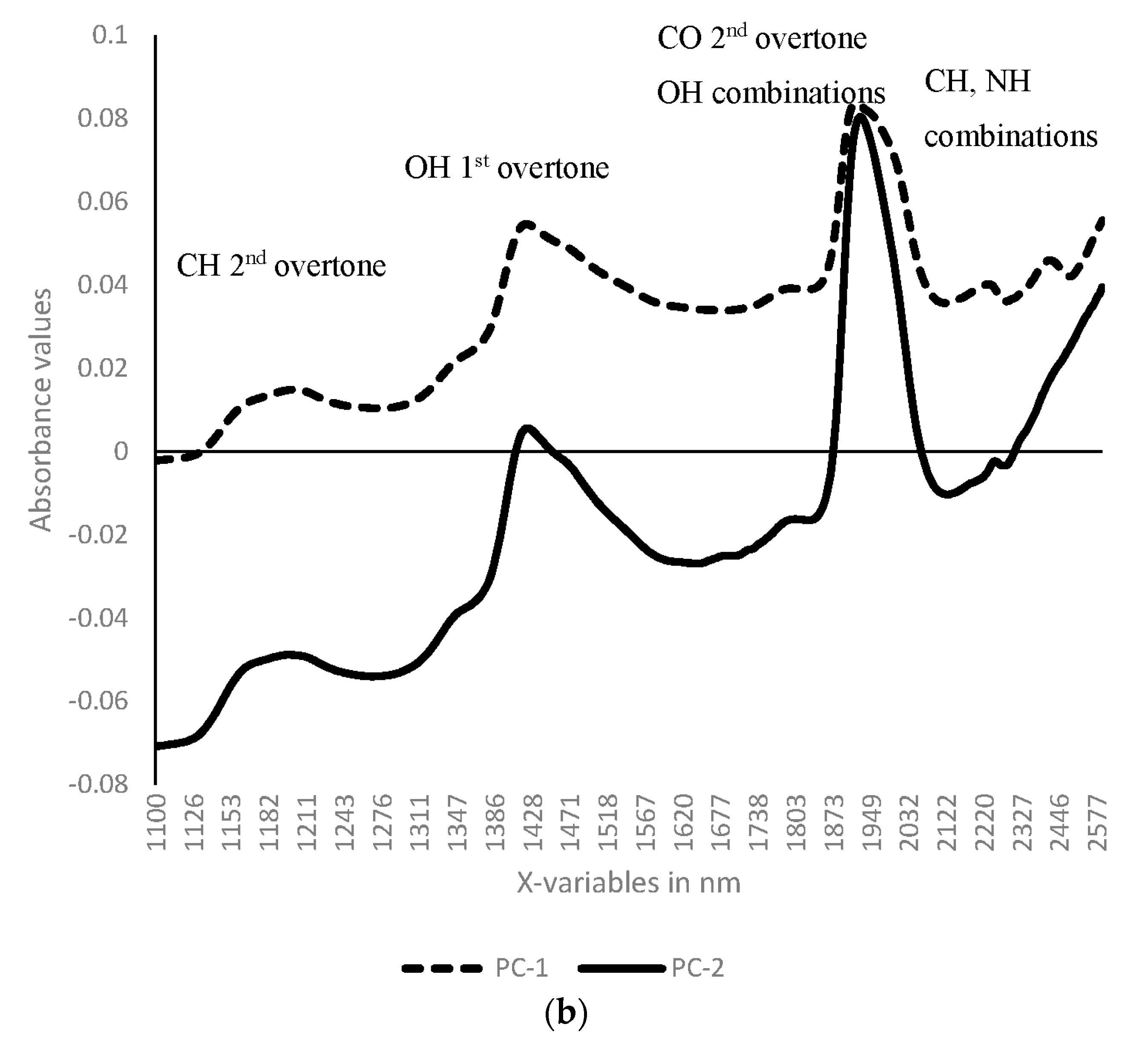
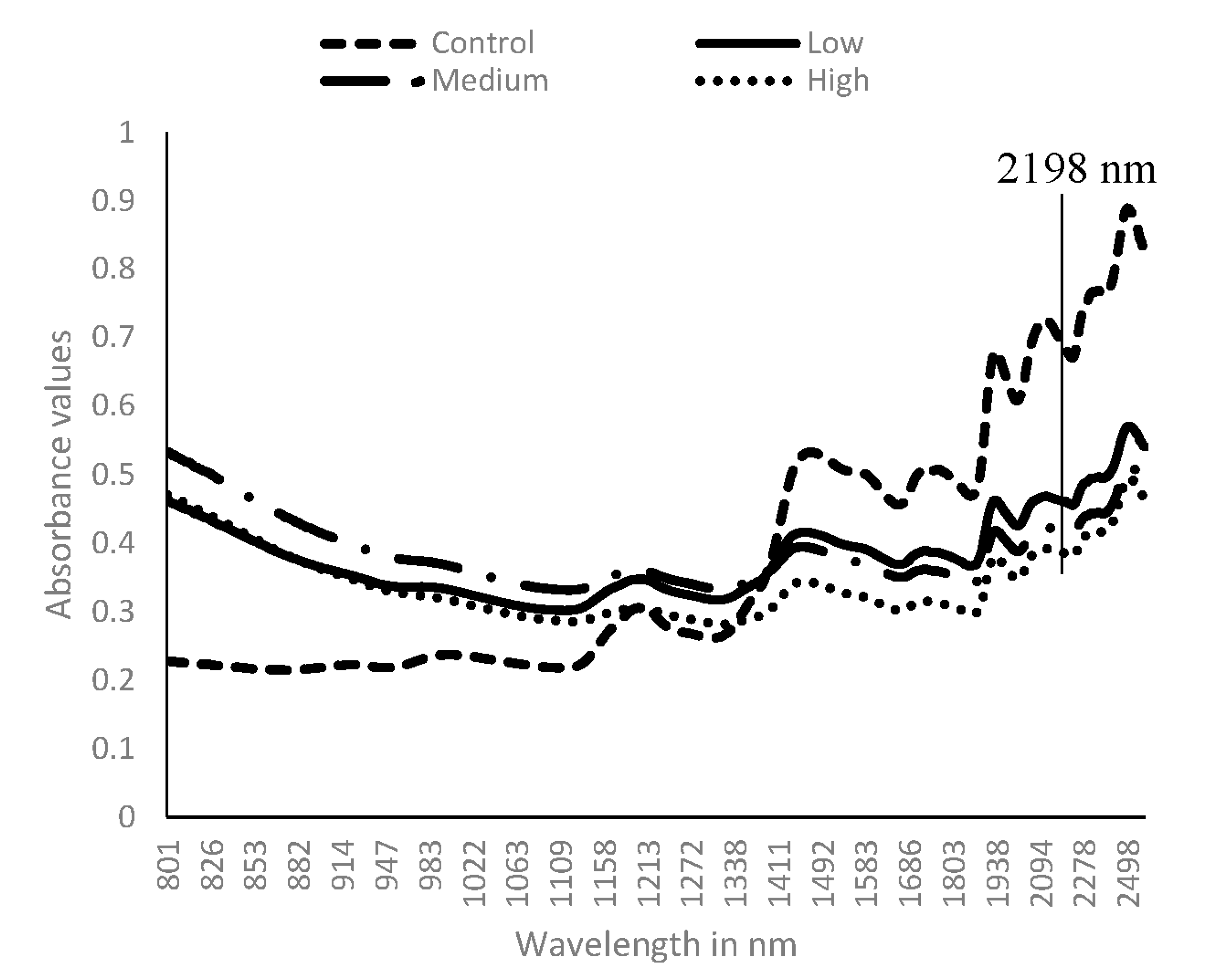
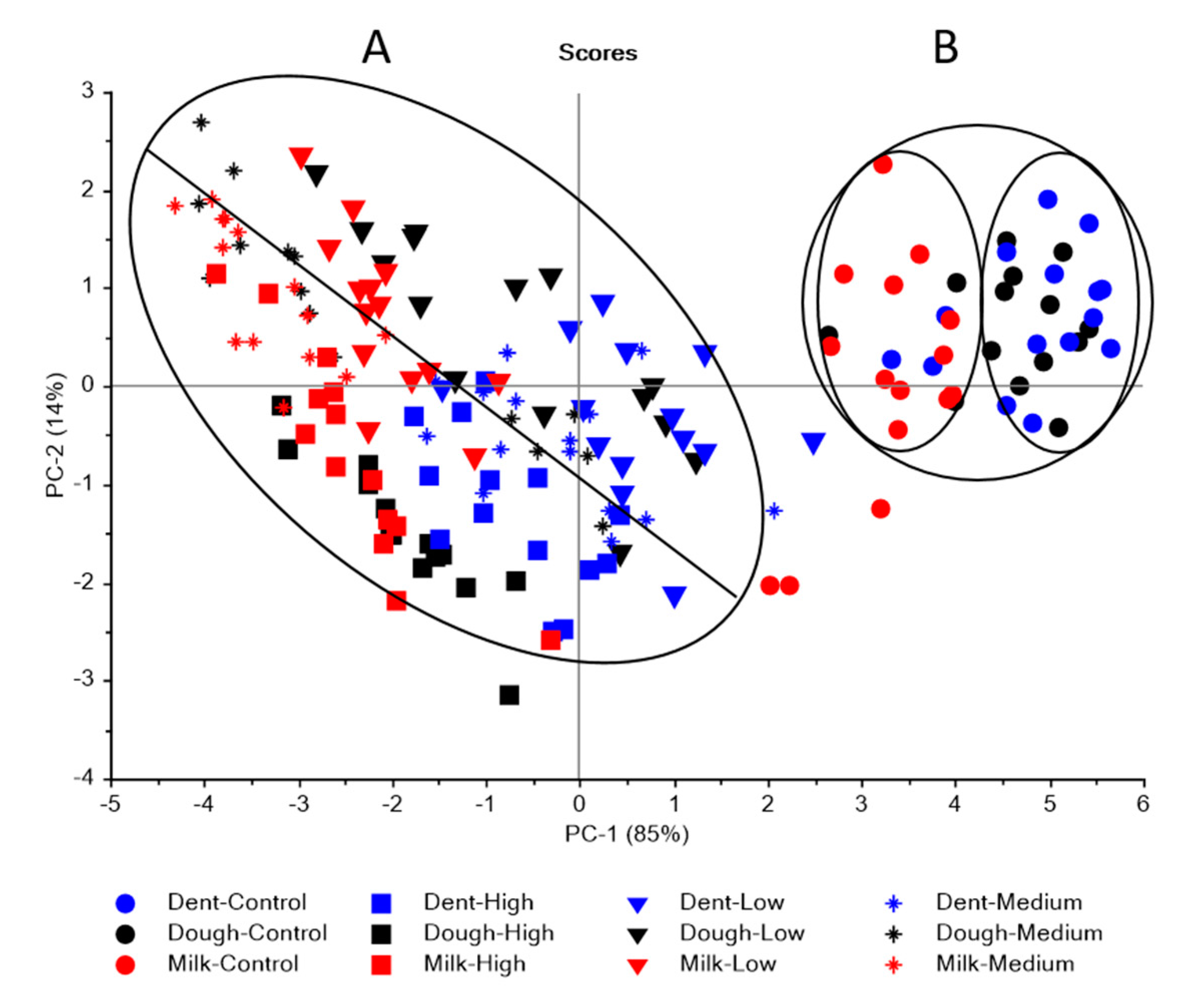
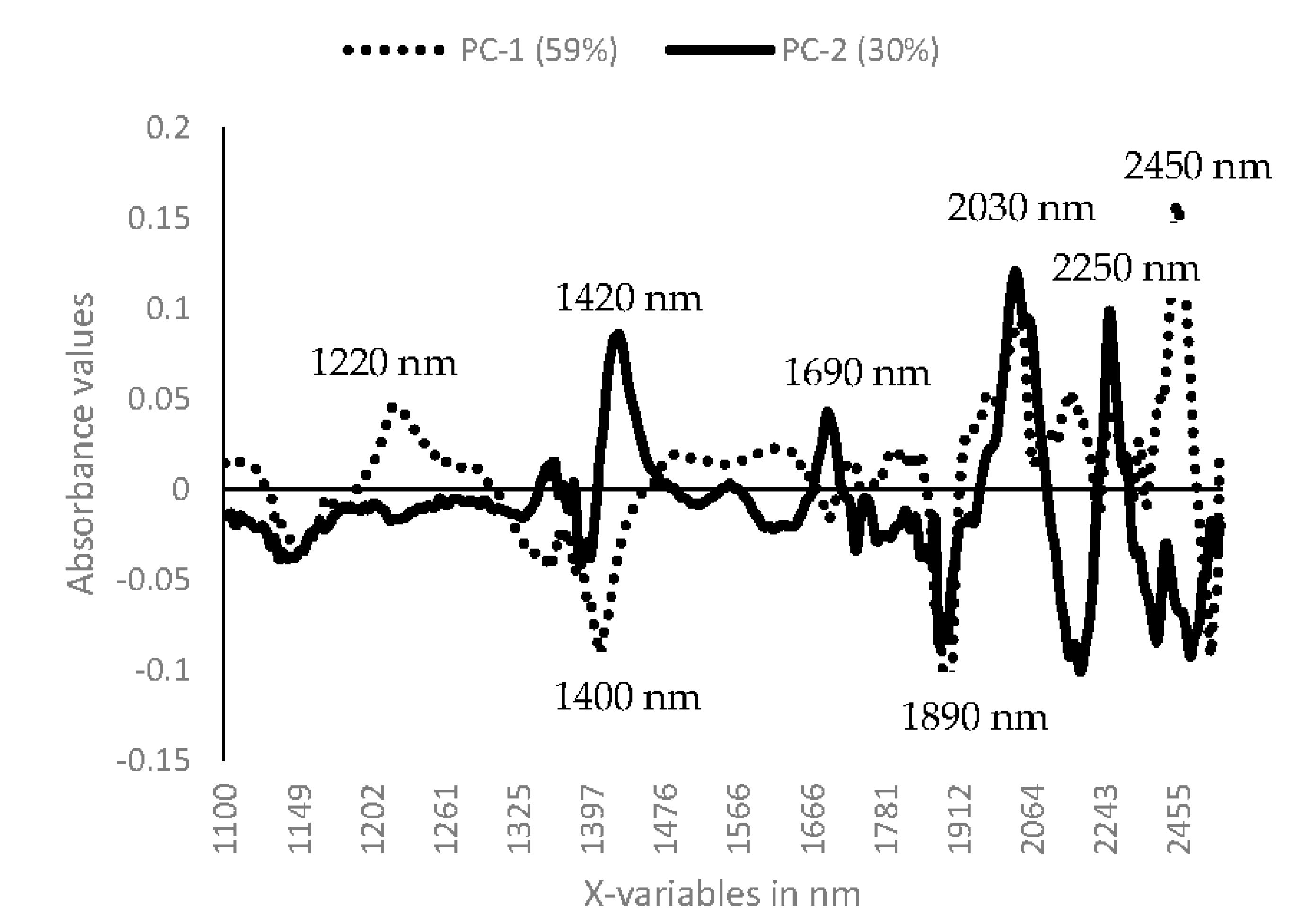
3.1. Near Infrared Spectral Data from Maize Kernels Prior to Fungal Inoculation
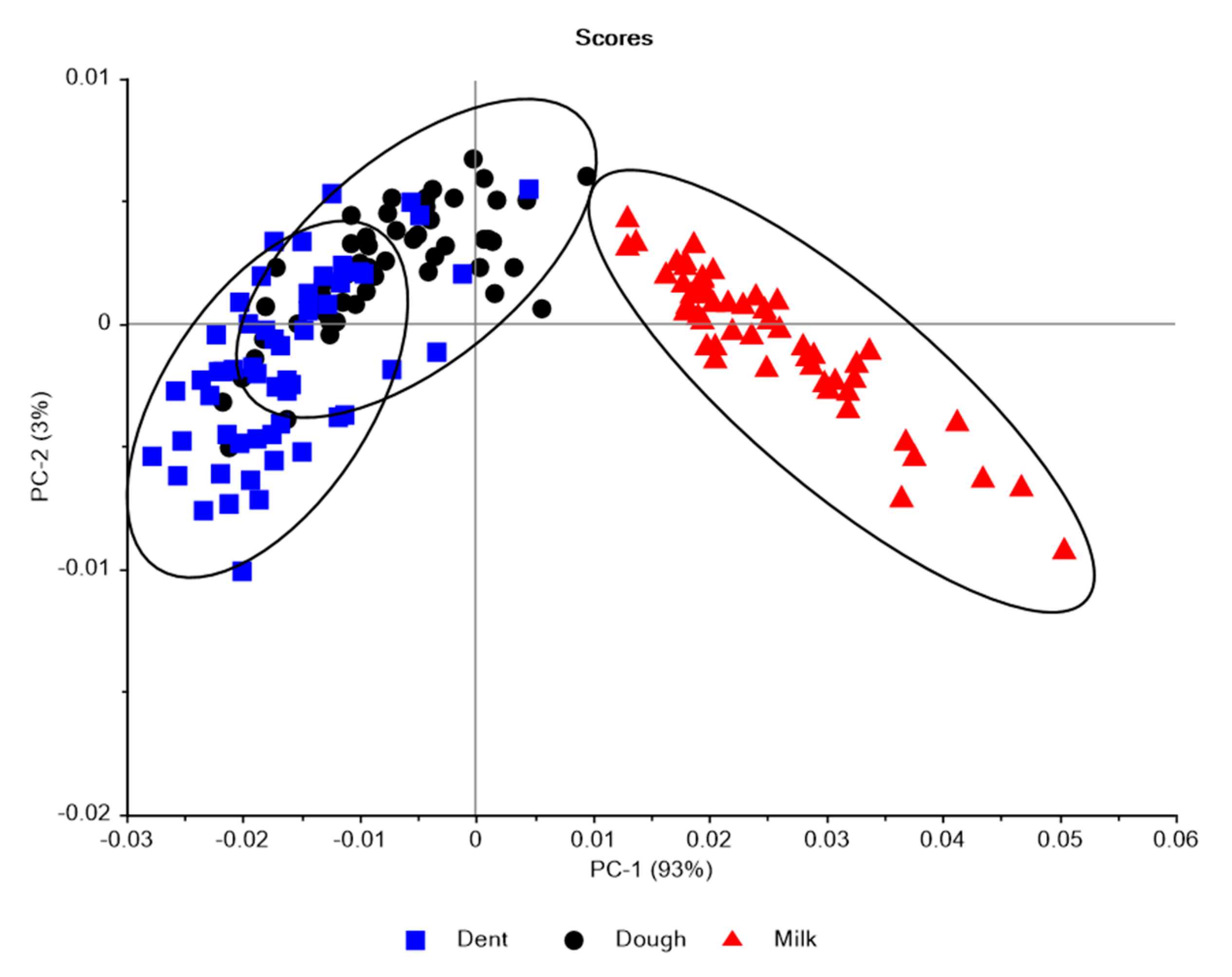
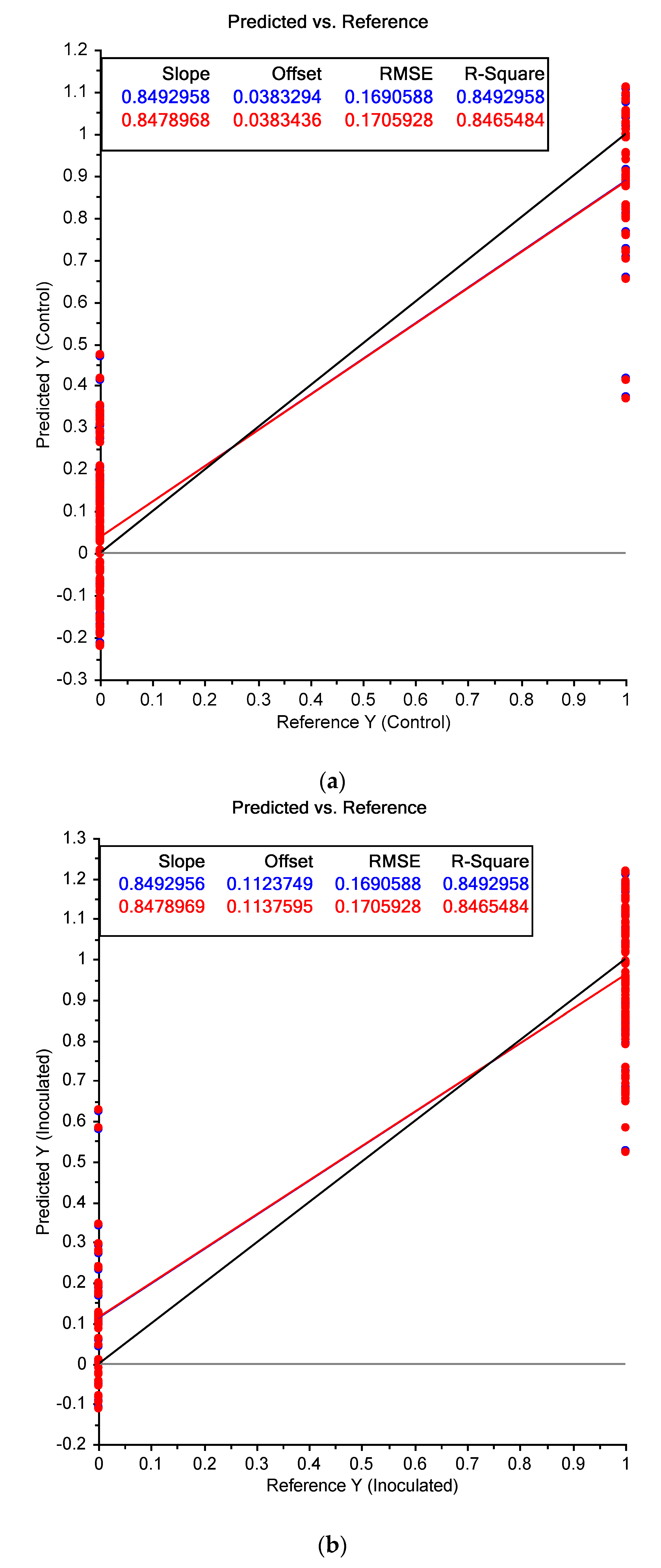
3.2. Near Infrared Spectral Data from Maize Kernels after Fungal Inoculation
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Smith, R.H.; McKernan, W. Hepatotoxic action of chromatographically separated fractions of Aspergillus flavus extracts. Nature 1962, 195, 1301–1303. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.M.; Sant’Ana, A.S.; Fungaro, M.H. P.; Silva, J.J.; do Nascimento, M.D.S.; Frisvad, J.C.; Taniwaki, M.H. The biodiversity of Aspergillus section Flavi and aflatoxins in the Brazilian peanut production chain. Food Res. Int. 2017, 94, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Krishnamachari, K.; Nagarajan, V.; Bhat, R.; Tilak, T. Hepatitis due to aflatoxicosis: An outbreak in western India. Lancet 1975, 305, 1061–1063. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Outbreak of aflatoxin poisoning—Eastern and central provinces, Kenya, January to July 2004. Morb. Mortal. Wkly. Rep. 2004, 53, 790–793. [Google Scholar]
- DPG Cholera Situation Updates and Response. Available online: http://www.tzdpg.or.tz/ (accessed on 6 February 2017).
- Udomkun, P.; Wiredu, A.N.; Nagle, M.; Müller, J.; Vanlauwe, B.; Bandyopadhyay, R. Innovative technologies to manage aflatoxins in foods and feeds and the profitability of application—A review. Food Control 2017, 76, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Weaver, M.A.; Abbas, H.K.; Jin, X.; Elliott, B. Efficacy of water-dispersible formulations of biological control strains of Aspergillus flavus for aflatoxin management in corn. Food Addit. Contam. Part A 2016, 33, 346–351. [Google Scholar]
- Battilani, P.; Leggieri, M.C. Predictive modelling of aflatoxin contamination to support maize chain management. World Mycotoxin J. 2015, 8, 161–170. [Google Scholar] [CrossRef]
- Williams, S.B.; Murdock, L.L.; Baributsa, D. Storage of maize in Purdue Improved Crop Storage (PICS) bags. PLoS ONE 2017, 12, e0168624. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lei, Y.; Wan, L.; Yan, L.; Lv, J.; Dai, X.; Ren, X.; Guo, W.; Jiang, H.; Liao, B. Comparative transcript profiling of resistant and susceptible peanut post-harvest seeds in response to aflatoxin production by Aspergillus flavus. BMC Plant Biol. 2016, 16, 54. [Google Scholar] [CrossRef] [PubMed]
- Manoza, F.S.; Mushongi, A.A.; Harvey, J.; Wainaina, J.; Wanjuki, I.; Ngeno, R.; Darnell, R.; Gnonlonfin, B.G.; Massomo, S.M. Potential of using host plant resistance, nitrogen and phosphorus fertilizers for reduction of Aspergillus flavus colonization and aflatoxin accumulation in maize in Tanzania. Crop Prot. 2017, 93, 98–105. [Google Scholar] [CrossRef]
- Arzandeh, S.; Jinap, S. Effect of initial aflatoxin concentration, heating time and roasting temperature on aflatoxin reduction in contaminated peanuts and process optimisation using response surface modelling. Int. J. Food Sci. Technol. 2011, 46, 485–491. [Google Scholar] [CrossRef]
- Yao, H.; Hruska, Z.; Di Mavungu, D. Developments in detection and determination of aflatoxins. World Mycotoxin J. 2015, 8, 181–191. [Google Scholar] [CrossRef]
- Cheli, F.; Battaglia, D.; Gallo, R.; Dell’Orto, V. EU legislation on cereal safety: An update with a focus on mycotoxins. Food Control 2014, 37, 315–325. [Google Scholar] [CrossRef]
- Torres, A.M.; Barros, G.G.; Palacios, S.A.; Chulze, S.N.; Battilani, P. Review on pre- and post-harvest management of peanuts to minimize aflatoxin contamination. Food Res. Int. 2014, 62, 11–19. [Google Scholar] [CrossRef]
- Bowers, E.; Hellmich, R.; Munkvold, G. Comparison of fumonisin contamination using HPLC and ELISA methods in bt and near-isogenic maize hybrids infested with European corn borer or western bean cutworm. J. Agric. Food Chem. 2014, 62, 6463–6472. [Google Scholar] [CrossRef] [PubMed]
- Dzuman, Z.; Zachariasova, M.; Veprikova, Z.; Godula, M.; Hajslova, J. Multi-analyte high performance liquid chromatography coupled to high resolution tandem mass spectrometry method for control of pesticide residues, mycotoxins, and pyrrolizidine alkaloids. Anal. Chim. Acta 2015, 863, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Nishimwe, K.; Wanjuki, I.; Karangwa, C.; Darnell, R.; Harvey, J. An initial characterization of aflatoxin B1 contamination of maize sold in the principal retail markets of Kigali, Rwanda. Food Control 2017, 73, 574–580. [Google Scholar] [CrossRef]
- Song, S.; Liu, N.; Zhao, Z.; Njumbe Ediage, E.; Wu, S.; Sun, C.; De Saeger, S.; Wu, A. Multiplex lateral flow immunoassay for mycotoxin determination. Anal. Chem. 2014, 86, 4995–5001. [Google Scholar] [CrossRef] [PubMed]
- Kandpal, L.M.; Lee, S.; Kim, M.S.; Bae, H.; Cho, B.-K. Short wave infrared (SWIR) hyperspectral imaging technique for examination of aflatoxin B1 (AFB1) on corn kernels. Food Control 2015, 51, 171–176. [Google Scholar] [CrossRef]
- Herschel, W. Experiments on the refrangibility of the invisible rays of the Sun. Philos. Trans. R. Soc. Lond. 1800, 90, 284–292. [Google Scholar] [CrossRef]
- Williams, P.; Norris, K. Near-Infrared Technology in the Agricultural and Food Industries; American Association of Cereal Chemists, Inc.: St. Paul, MN, USA, 1987. [Google Scholar]
- Finney, E., Jr.; Norris, K. Determination of moisture in corn kernels by near-infrared transmittance measurements. Trans. Am. Soc. Agric. Eng. 1978, 21, 581–584. [Google Scholar] [CrossRef]
- Hruschka, W.R.; Norris, K.H. Least-squares curve fitting of near infrared spectra predicts protein and moisture content of ground wheat. Appl. Spectrosc. 1982, 36, 261–265. [Google Scholar] [CrossRef]
- Williams, P.C.; Norris, K.H.; Sobering, D.C. Determination of protein and moisture in wheat and barley by near-infrared transmission. J. Agric. Food Chem. 1985, 33, 239–244. [Google Scholar] [CrossRef]
- Durmuş, E.; Güneş, A.; Kalkan, H. Detection of aflatoxin and surface mould contaminated figs by using Fourier-transform near-infrared (FT-NIR) reflectance spectroscopy. J. Sci. Food Agric. 2016, 97, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Qiang, Z.; Fuguo, J.; Chenghai, L.; Jingkun, S.; Xianzhe, Z. Rapid detection of aflatoxin B1 in paddy rice as analytical quality assessment by near infrared spectroscopy. Int. J. Agric. Biol. Eng. 2014, 7, 127–133. [Google Scholar]
- Wang, W.; Lawrence, K.C.; Ni, X.; Yoon, S.-C.; Heitschmidt, G.W.; Feldner, P. Near-infrared hyperspectral imaging for detecting aflatoxin B1 of maize kernels. Food Control 2015, 51, 347–355. [Google Scholar] [CrossRef]
- Qiao, X.; Jiang, J.; Qi, X.; Guo, H.; Yuan, D. Utilization of spectral-spatial characteristics in shortwave infrared hyperspectral images to classify and identify fungi-contaminated peanuts. Food Chem. 2017, 220, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Yao, H.; Hruska, Z.; Kincaid, R.; Brown, R.; Bhatnagar, D.; Cleveland, T. Classification of corn kernels contaminated with aflatoxins using fluorescence and reflectance hyperspectral images analysis. In SPIE Sensing Technology + Applications; International Society for Optics and Photonics: Baltimore, MD, USA, 2015; p. 94880M. [Google Scholar]
- Liang, P.-S.; Slaughter, D.C.; Ortega-Beltran, A.; Michailides, T.J. Detection of fungal infection in almond kernels using near-infrared reflectance spectroscopy. Biosyst. Eng. 2015, 137, 64–72. [Google Scholar] [CrossRef]
- Yao, H.; Hruska, Z.; Kincaid, R.; Brown, R.L.; Bhatnagar, D.; Cleveland, T.E. Detecting maize inoculated with toxigenic and atoxigenic fungal strains with fluorescence hyperspectral imagery. Biosyst. Eng. 2013, 115, 125–135. [Google Scholar] [CrossRef]
- Lee, K.-M.; Davis, J.; Herrman, T.J.; Murray, S.C.; Deng, Y. An empirical evaluation of three vibrational spectroscopic methods for detection of aflatoxins in maize. Food Chem. 2015, 173, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ibañez, V.; Soldado, A.; Martínez-Fernández, A.; de la Roza-Delgado, B. Application of near infrared spectroscopy for rapid detection of aflatoxin B1 in maize and barley as analytical quality assessment. Food Chem. 2009, 113, 629–634. [Google Scholar] [CrossRef]
- Reese, B.N.; Payne, G.A.; Nielsen, D.M.; Woloshuk, C.P. Gene expression profile and response to maize kernels by Aspergillus flavus. Phytopathology 2011, 101, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Dolezal, A.L.; Shu, X.; OBrian, G.R.; Nielsen, D.M.; Woloshuk, C.P.; Boston, R.S.; Payne, G.A. Aspergillus flavus infection induces transcriptional and physical changes in developing maize kernels. Front. Microbiol. 2014, 5, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.L. Grain Fill Stages in Corn. Available online: https://www.agry.purdue.edu/ext/corn/news/timeless/GrainFill.html (accessed on 6 February 2017).
- Baye, T.M.; Pearson, T.C.; Settles, A.M. Development of a calibration to predict maize seed composition using single kernel near infrared spectroscopy. J. Cereal Sci. 2006, 43, 236–243. [Google Scholar] [CrossRef]
- Cogdill, R.P.; Hurburgh, C.R. J.; Rippke, G.R.; Bajic, S.J.; Jones, R.W.; McClelland, J.F.; Jensen, T.C.; Liu, J. Single-kernel maize analysis by near-infrared hyperspectral imaging. Trans. Am. Soc. Agric. Eng. 2004, 47, 311–320. [Google Scholar] [CrossRef]
- An, D.; Cui, Y.; Liu, X.; Jia, S.; Zheng, S.; Che, X.; Liu, Z.; Zhang, X.; Zhu, D.; Li, S. Effects of varieties, producing areas, ears, and ear positions of single maize kernels on near-infrared spectra for identification and traceability. PLoS ONE 2016, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Swarbrick, B. Chemometric techniques for robust calibration development. In Proceedings of the 16th Australian NIR Spectroscopy Conference, Coffs Harbour, Australia, 4–7 May 2014; Australian Near Infrared Spectroscopy Group: Coffs Harbour, NSW, Australia, 2014. [Google Scholar]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Growth characteristics of three Fusarium species evaluated by near-infrared hyperspectral imaging and multivariate image analysis. Appl. Microbiol. Biotechnol. 2012, 96, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, W.; Chu, X.; Li, C.; Kimuli, D. Early detection of Aspergillus parasiticus infection in maize kernels using near-infrared hyperspectral imaging and multivariate data analysis. Appl. Sci. 2017, 7, 90. [Google Scholar] [CrossRef]
- Dahm, D.J.; Dahm, K.D. The physics of near-infrared scattering. In Near-Infrared Technology in the Agricultural and Food Industries; Williams, P.C., Norris, K.H., Eds.; American Association of Cereal Chemists: St Paul, MN, USA, 2001; pp. 1–19. [Google Scholar]
- Reich, G. Near-infrared spectroscopy and imaging: Basic principles and pharmaceutical applications. Adv. Drug Deliv. Rev. 2005, 57, 1109–1143. [Google Scholar] [CrossRef] [PubMed]
- Manley, M. Near-infrared spectroscopy and hyperspectral imaging: Non-destructive analysis of biological materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef] [PubMed]
- Barnes, R.; Dhanoa, M.; Lister, S. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Wang, X.; Larkins, B.A. Genetic analysis of amino acid accumulation in opaque-2 maize endospoerm. Plant Physiol. 2001, 125, 1766–1777. [Google Scholar] [CrossRef] [PubMed]
- Afuakwa, J.J.; Crookston, K.R. Using the kernel milk line to visually monitor grain maturity in maize. Crop Sci. 1983, 24, 687–691. [Google Scholar] [CrossRef]
- Li, L.; Blanco, M.; Jane, J. Physicochemical properties of endosperm and pericarp starches during maize development. Carbohydr. Polym. 2007, 67, 630–639. [Google Scholar] [CrossRef]
- Manley, M.; Williams, P.; Nilsson, D.; Geladi, P. Near infrared hyperspectral imaging for the evaluation of endosperm texture in whole yellow maize (Zea maize L.) kernels. J. Agric. Food Chem. 2009, 57, 8761–8769. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falade, T.D.O.; Sultanbawa, Y.; Fletcher, M.T.; Fox, G. Near Infrared Spectrometry for Rapid Non-Invasive Modelling of Aspergillus-Contaminated Maturing Kernels of Maize (Zea mays L.). Agriculture 2017, 7, 77. https://doi.org/10.3390/agriculture7090077
Falade TDO, Sultanbawa Y, Fletcher MT, Fox G. Near Infrared Spectrometry for Rapid Non-Invasive Modelling of Aspergillus-Contaminated Maturing Kernels of Maize (Zea mays L.). Agriculture. 2017; 7(9):77. https://doi.org/10.3390/agriculture7090077
Chicago/Turabian StyleFalade, Titilayo D.O., Yasmina Sultanbawa, Mary T. Fletcher, and Glen Fox. 2017. "Near Infrared Spectrometry for Rapid Non-Invasive Modelling of Aspergillus-Contaminated Maturing Kernels of Maize (Zea mays L.)" Agriculture 7, no. 9: 77. https://doi.org/10.3390/agriculture7090077





