Genetic Engineering of Energy Crops to Reduce Recalcitrance and Enhance Biomass Digestibility
Abstract
:1. Introduction
2. Biomass Bioconversion and the Plant Cell Wall Complex
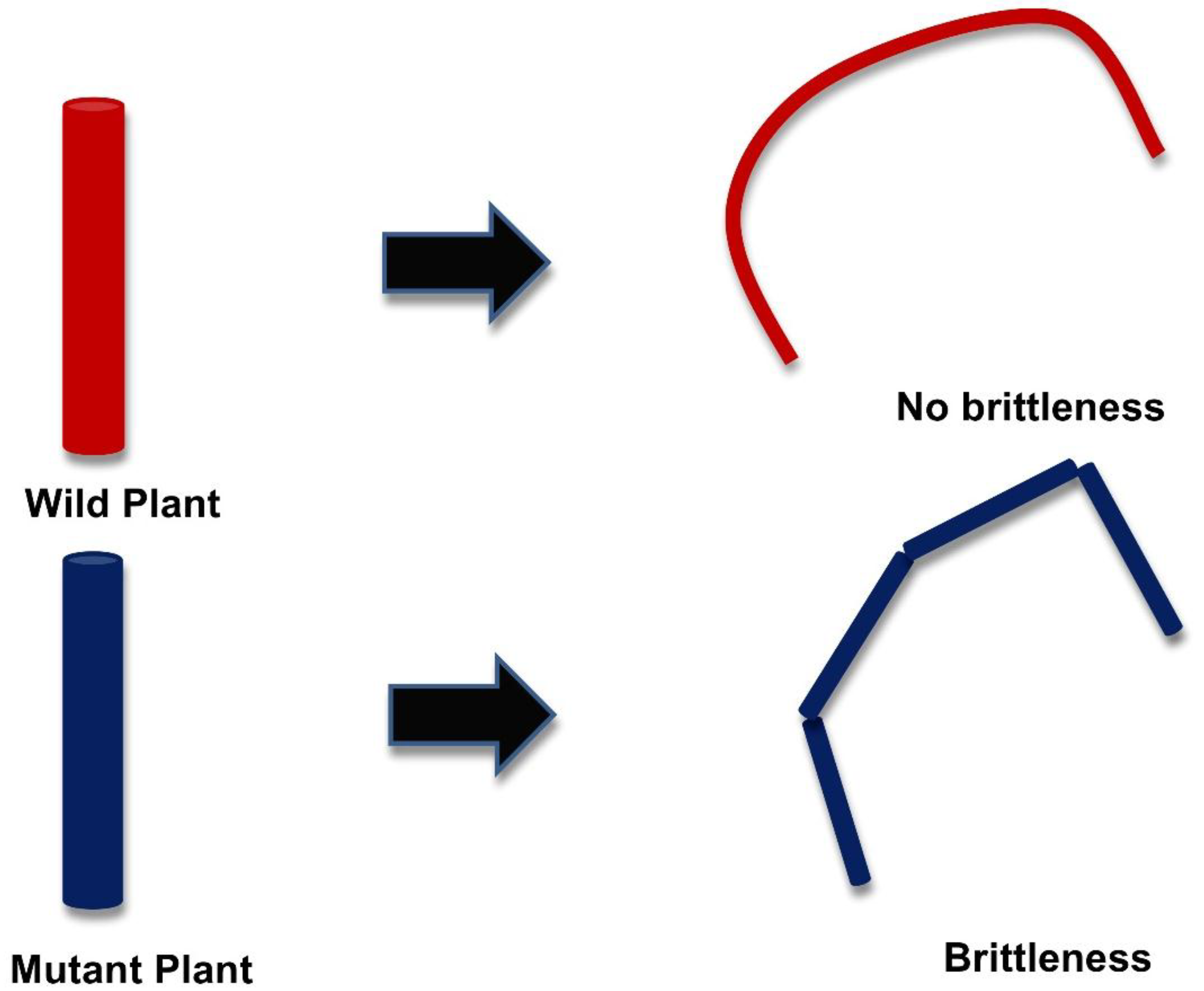
3. Genetic Modification in the Plant Cell Wall
3.1. Wall Polymer Biosynthesis
3.2. Wall Polymer Degradation
3.3. Cell Wall Network Construction
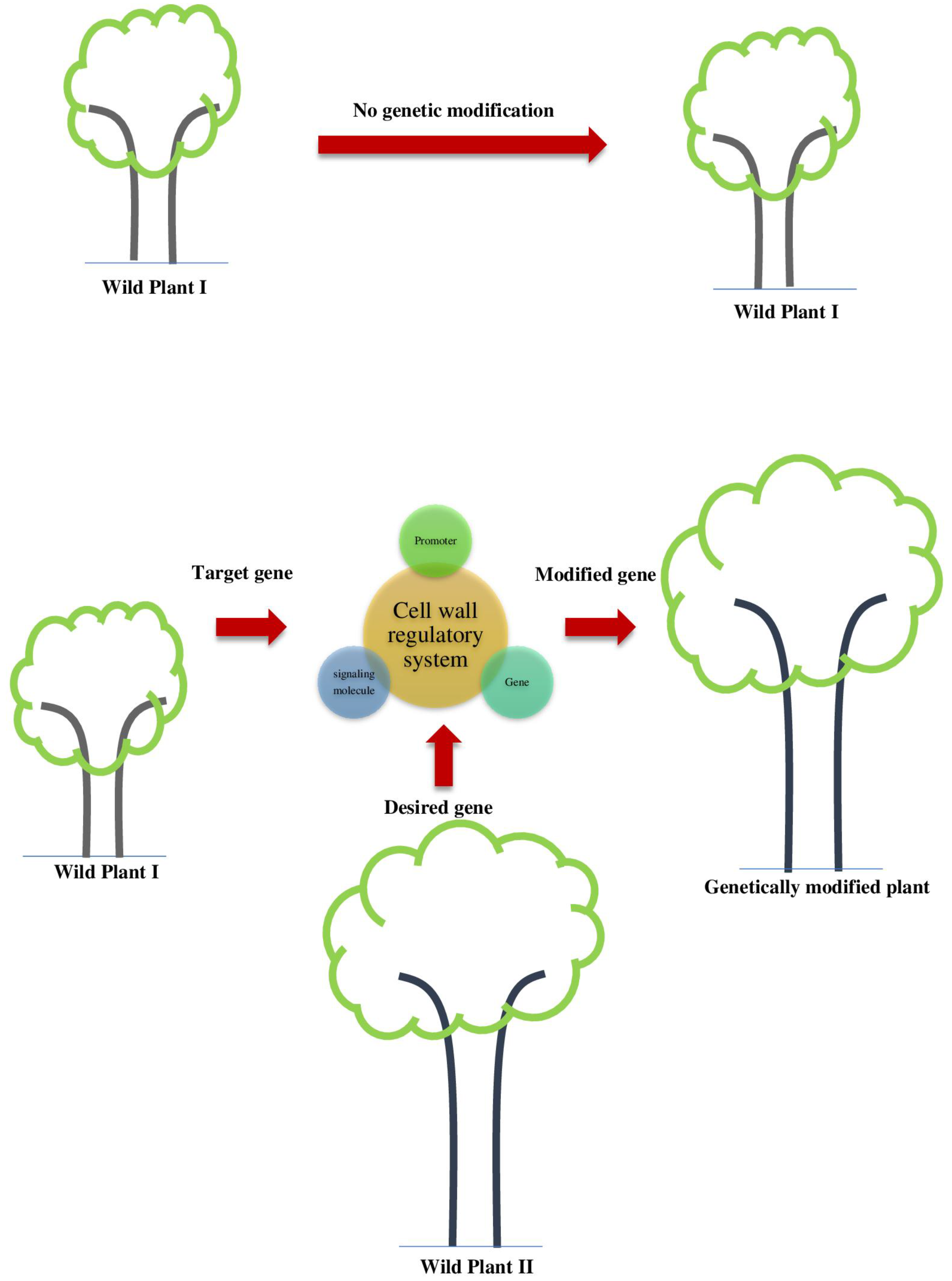
4. Approaches for Genetic Manipulations
5. Conclusions and Future Aspects
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Himmel, M.E.; Bayer, E.A. Lignocellulose conversion to biofuels: Current challenges, global perspectives. Curr. Opin. Biotechnol. 2009, 20, 316–317. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Peng, L.C. The diversity of lignocellulosic biomass resources and their evaluation for use as biofuels and chemicals. In Biological Conversion of Biomass for Fuels and Chemicals: Explorations from Natural Biomass Utilization Systems; Sun, J.Z., Ding, S.Y., Peterson, J.D., Eds.; Royal Society of Chemistry: Cambridge, UK, 2013; pp. 83–109. [Google Scholar]
- Wang, Y.T.; Xu, Z.D.; Peng, L.C. Model of plant cell wall ditches structure for enhancing biomass saccharification and plant resistances. Sci. Sin. Vitae 2014, 44, 766–774. [Google Scholar] [CrossRef]
- Xie, G.S.; Peng, L.C. Genetic engineering of energy crops: A strategyfor biofuel production in China. J. Integr. Plant Biol. 2011, 53, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Li, F.C.; Zhang, M.L.; Guo, K.; Hu, Z.; Zhang, R.; Feng, Y.Q.; Yi, X.; Zou, W.; Wang, L.; Wu, C.; et al. High-level hemicellulosic arabinose predominately affects lignocellulose crystallinity for genetically enhancing both plant lodging resistance and biomass enzymatic digestibility in rice mutants. Plant Biotechnol. J. 2015, 13, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Glass, M.; Barkwill, S.; Unda, F.; Mansfield, S.D. Endo-beta-1,4-glucanases impact plant cell wall development by influencing cellulose crystallization. J. Integr. Plant Biol. 2015, 57, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.W.; Vega-Sánchez, M.; Verhertbruggen, Y.; Chiniquy, D.; Canlas, P.E.; Fagerström, A.; Prak, L.; Christensen, U.; Oikawa, A.; Chern, M.; et al. Inactivation of OsIRX10 leads to decreased xylan content in rice culm cell walls and improved biomass saccharification. Mol. Plant 2013, 6, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.X.; Teng, Q.; Zhong, R.Q.; Ye, Z.H. The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol. 2013, 54, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.P.; Emsley, L. The 2D MAS NMR spin-echo experiment: The determination of 13C–13C J couplings in a solid-state cellulose sample. J. Magn. Reson. 2004, 171, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Kaida, R.; Kaku, T.; Baba, K.; Oyadomari, M.; Watanabe, T.; Nishida, K.; Kanaya, T.; Shani, Z.; Shoseyov, O.; Hayashi, T. Loosening xyloglucan accelerates the enzymatic degradation of cellulose in wood. Mol. Plant 2009, 2, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Girio, F.M.; Fonseca, C.; Carvalheiro, F.; Duarte, L.C.; Marques, S.; Bogel-Łukasik, R. Hemicelluloses for fuel ethanol: A review. Bioresour. Technol. 2010, 101, 4775–4800. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Paritosh, K.; Pareek, N.; Chawade, A.; Vivekanand, V. De-construction of major Indian cereal crop residues through chemical pretreatment for improved biogas production: An overview. Renew. Sustain. Energy Rev. 2018, 90, 160–170. [Google Scholar] [CrossRef]
- Chawade, A.; Alexandersson, E.; Bengtsson, T.; Andreasson, E.; Levander, F. Targeted Proteomics Approach for Precision Plant Breeding. J. Proteome Res. 2016, 15, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Lindlof, A.; Brautigam, M.; Chawade, A.; Olsson, O.; Olsson, B. In silico analysis of promoter regions from cold-induced genes in rice (Oryza sativa L.) and Arabidopsis thaliana reveals the importance of combinatorial control. Bioinformatics 2009, 25, 1345–1348. [Google Scholar] [CrossRef] [PubMed]
- Chawade, A.; Sikora, P.; Bräutigam, M.; Larsson, M.; Vivekanand, V.; Nakash, M.; Chen, T.; Olsson, O. Development and characterization of an oat TILLING-population and identification of mutations in lignin and β-glucan biosynthesis genes. BMC Plant Biol. 2010, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Vivekanand, V.; Chawade, A.; Larsson, M.; Larsson, A.; Olsson, O. Identification and qualitative characterization of high and low lignin lines from an oat TILLING population. Ind. Crops Prod. 2014, 59, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Guo, K.; Zou, W.; Feng, Y.; Zhang, M.; Zhang, J.; Tu, F.; Xie, G.; Wang, L.; Wang, Y.; Klie, S.; et al. An integrated genomic and metabolomic frame work for cell wall biology in rice. BMC Genom. 2014, 15, 596. [Google Scholar] [CrossRef] [PubMed]
- Brabham, C.; DeBolt, S. Chemical genetics to examine cellulose biosynthesis. Front. Plant Sci. 2013, 3, 309. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.C.; Kawagoe, Y.S.; Hogan, P.; Delmer, D. Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 2002, 295, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Xia, T.; Xu, W.; Chen, T.T.; Li, X.L.; Fan, J.; Wang, R.; Feng, S.; Wang, Y.; Wang, B.; et al. An integrative analysis of four CESA isoforms specific for fiber cellulose production between Gossypium hirsutum and Gossypium barbadense. Planta 2013, 237, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.T.; Shirley, N.J.; Singh, R.R.; Henderson, M.; Dhugga, K.S.; Mayo, G.M.; Fincher, G.B.; Burton, R.A. Powerful regulatory systems and post-transcriptional gene silencing resist increases in cellulose content in cell walls of barley. BMC Plant Biol. 2015, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Song, X.Q.; Liu, L.F.; Jiang, Y.J.; Zhang, B.C.; Gao, Y.P.; Liu, X.L.; Lin, Q.S.; Ling, H.Q.; Zhou, Y.H. Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants. Mol. Plant 2013, 6, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Biswal, A.K.; Hao, Z.Y.; Pattathil, S.; Yang, X.H.; Winkeler, K.; Collins, C.; Mohanty, S.S.; Richardson, E.A.; Gelineo-Albersheim, I.; Hunt, K.; et al. Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnol. Biofuels 2015, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Sattler, S.E.; Palmer, N.A.; Saballos, A.; Greene, A.M.; Xin, Z.G.; Sarath, G.; Vermerris, W.; Pedersen, J.F. Identification and characterization of four missense mutations in Brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. Bioenergy Res. 2012, 5, 855–865. [Google Scholar] [CrossRef]
- Xu, B.; Escamilla-Treviño, L.L.; Sathitsuksanoh, N.; Shen, Z.; Shen, H.; Zhang, Y.H.; Dixon, R.A.; Zhao, B. Silencing of 4-coumarate: Coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 2011, 192, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Francocci, F.; Ferrari, S.; Volpi, C.; Bellincampi, D.; Galletti, R.; D’Ovidio, R.; De Lorenzo, G.; Cervone, F. Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc. Natl. Acad. Sci. USA 2010, 107, 616–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswal, A.K.; Soeno, K.; Gandla, M.L.; Immerzeel, P.; Pattathil, S.; Lucenius, J.; Serimaa, R.; Hahn, M.G.; Moritz, T.; Jönsson, L.J.; et al. Aspen pectate lyase PtxtPL1-27 mobilizes matrix polysaccharides from woody tissues and improves saccharification yield. Biotechnol. Biofuels 2014, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, P.; Saha, P.; RayT, T.Y.H.; Yang, D.; Cannon, M.C. EXTENSIN18 is required for full male fertility as well as normal vegetative growth in Arabidopsis. Front. Plant Sci. 2015, 6, 553. [Google Scholar] [PubMed]
- Tavares, E.Q.P.; De Souza, A.P.; Buckeridge, M.S. How endogenous plant cell-wall degradation mechanisms can help achieve higher efficiency in saccharification of biomass. J. Exp. Bot. 2015, 66, 4133–4143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Souza, A.P.; Grandis, A.; Leite, D.C.C.; Buckeridge, M.S. Sugarcane as a bioenergy source: History, performance, and perspectives for second-generation bioethanol. BioEnergy Res. 2014, 7, 24–35. [Google Scholar] [CrossRef]
- Derba-Maceluch, M.; Awano, T.; Takahashi, J.; Lucenius, J.; Ratke, C.; Kontro, I.; Busse-Wicher, M.; Kosik, O.; Tanaka, R.; Winzéll, A.; et al. Suppression of xylan endotransglycosylase PtxtXyn10A affects cellulose microfibril angle in secondary wall in aspen wood. New Phytol. 2014, 205, 666–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomassetti, S.; Pontiggia, D.; Verrascina, I.; Reca, I.B.; Francocci, F.; Salvi, G.; Cervone, F.; Ferrari, S. Controlled expression of pectic enzymes in Arabidopsis thaliana enhances biomass conversion without adverse effects on growth. Phytochemistry 2015, 112, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.Q.; Ye, Z.H. Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 2007, 10, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Petersen, P.D.; Lau, J.; Ebert, B.; Yang, F.; Verhertbruggen, Y.; Kim, J.S.; Varanasi, P.; Suttangkakul, A.; Auer, M.; Loqué, D.; et al. Engineering of plants with improved properties as biofuels feedstocks by vesselspecific complementation of xylan biosynthesis mutants. Biotechnol. Biofuels 2012, 5, 84. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sakamoto, S.; Kawai, T.; Kobayashi, Y.; Sato, K.; Ichinose, Y.; Yaoi, K.; Akiyoshi-Endo, M.; Sato, H.; Takamizo, T.; et al. Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front. Plant Sci. 2013, 4, 383. [Google Scholar] [CrossRef] [PubMed]
- Ralph, J.; Akiyama, T.; Kim, H.; Lu, F.; Schatz, P.F.; Marita, J.M.; Ralph, S.A.; Reddy, M.S.; Chen, F.; Dixon, R.A. Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J. Biol. Chem. 2006, 281, 8843–8853. [Google Scholar] [CrossRef] [PubMed]
- Wilkerson, C.G.; Mansfield, S.D.; Lu, F.; Withers, S.; Park, J.Y.; Karlen, S.D.; Gonzales-Vigil, E.; Padmakshan, D.; Unda, F.; Rencoret, J.; et al. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 2014, 344, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Abramson, M.; Shoseyov, O.; Shani, Z. Plant cell wall reconstruction toward improved lignocellulosic production and processability. Plant Sci. 2010, 178, 61–72. [Google Scholar] [CrossRef]
- Vega-Sánchez, M.E.; Loqué, D.; Lao, J.; Catena, M.; Verhertbruggen, Y.; Herter, T.; Yang, F.; Harholt, J.; Ebert, B.; Baidoo, E.E.; et al. Engineering temporal accumulation of a low recalcitrance polysaccharide leads to increased C6 sugar content in plant cell walls. Plant Biotechnol. J. 2015, 13, 903–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, D.M.; Corbin, K.; Wang, T.; Gutierrez, R.; Bertolo, A.L.; Petti, C.; Smilgies, D.M.; Estevez, J.M.; Bonetta, D.; Urbanowicz, B.R.; et al. Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc. Natl. Acad. Sci. USA 2012, 109, 4098–4103. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, D.K.; Stork, J.; DeBolt, S.; Maiti, I.B. Manipulating cellulose biosynthesis by expression of mutant Arabidopsis proM24: CESA3ixr1-2 gene in transgenic tobacco. Plant Biotechnol. J. 2013, 11, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Handakumbura, P.P.; Matos, D.A.; Osmont, K.S.; Harrington, M.J.; Heo, K.; Kafle, K.; Kim, S.H.; Baskin, T.I.; Hazen, S.P. Perturbation of Brachypodium distachyon CELLULOSE SYNTHASE A4 or 7 results in abnormal cell walls. BMC Plant Biol. 2013, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Li, D.Y.; Liu, X.; Ji, C.J.; Hao, L.L.; Zhao, X.F.; Li, X.B.; Chen, C.Y.; Cheng, Z.K.; Zhu, L.H. OsMYB103L, an R2R3MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol. 2014, 14, 158. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wang, S.; Zhang, B.; Shang-Guan, K.; Shi, Y.; Zhang, D.; Liu, X.; Wu, K.; Xu, Z.; Fu, X.; et al. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 2015, 27, 1681–1696. [Google Scholar] [CrossRef] [PubMed]
- Chiniquy, D.; Sharma, V.; Schultink, A.; Baidoo, E.E.; Rautengarten, C.; Cheng, K.; Carroll, A.; Ulvskov, P.; Harholt, J.; Keasling, J.D.; et al. XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc. Natl. Acad. Sci. USA 2012, 109, 17117–17122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawar, P.M.A.; Derba-Maceluch, M.; Chong, S.L.; Gómez, L.D.; Miedes, E.; Banasiak, A.; Ratke, C.; Gaertner, C.; Mouille, G.; McQueen-Mason, S.J.; et al. Expression of fungal acetyl xylan esterase in Arabidopsis thaliana improves saccharification of stem lignocellulose. Plant Biotechnol. J. 2015, 14, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.; Rudsander, U.J.; Hedenstrom, M.; Banasiak, A.; Harholt, J.; Amelot, N.; Immerzeel, P.; Ryden, P.; Endo, S.; Ibatullin, F.M.; et al. KORRIGAN1 and its aspen homolog PttCel9A1 decrease cellulose crystallinity in Arabidopsis stems. Plant Cell Physiol. 2009, 50, 1099–1115. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.Y.; Cheng, K.; Pauly, M. Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol. Plant 2013, 6, 1373–1375. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.Z.; Peña, M.J.; Renna, L.; Avci, U.; Pattathil, S.; Tuomivaara, S.T.; Li, X.; Reiter, W.D.; Brandizzi, F.; Hahn, M.G.; et al. Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol. 2015, 167, 1296–1306. [Google Scholar] [CrossRef] [PubMed]
- Schultink, A.; Naylor, D.; Dama, M.; Pauly, M. The role of the plant-specific ALTERED XYLOGLUCAN9 protein in Arabidopsis cell wall polysaccharide O-acetylation. Plant Physiol. 2015, 167, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Giraldo, L.; Bhattarai, K.; Pislariu, C.; Nakashima, J.; Jikumaru, Y.; Kamiya, Y.; Udvardi, M.K.; Monteros, M.J.; Dixon, R.A. Lignin modification leads to increased nodule numbers in alfalfa. Plant Physiol. 2014, 164, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl Shikimate Esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Bouvier d’Yvoire, M.; Bouchabke-Coussa, O.; Voorend, W.; Antelme, S.; Cézard, L.; Legée, F.; Lebris, P.; Legay, S.; Whitehead, C.; McQueen-Mason, S.J.; et al. Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J. 2013, 73, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, R.; Lepléc, J.C.; Aerts, D.; Stormea, V.; Goeminne, G.; Ivens, B.; Légée, F.; Lapierre, C.; Piens, K.; Van Montagu, M.C.; et al. Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. USA 2014, 111, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Fouad, W.M.; Vermerris, W.; Gallo, M.; Altpeter, F. RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol. J. 2012, 10, 1067–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, H.L.; Mazarei, M.; Labbe, N.; Kline, L.M.; Cheng, Q.; Windham, M.T.; Mann, D.G.; Fu, C.; Ziebell, A.; Sykes, R.W.; et al. Two-year field analysis of reduced recalcitrance transgenic switchgrass. Plant Biotechnol. J. 2014, 12, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Kim, J.I.; Tobimatsu, Y.; Ciesielski, P.N.; Anderson, N.A.; Ximenes, E.; Maeda, J.; Ralph, J.; Donohoe, B.S.; Ladisch, M.; et al. Disruption of mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 2014, 509, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Francocci, F.; Bastianelli, E.; Lionetti, V.; Ferrari, S.; De Lorenzo, G.; Bellincampi, D.; Cervone, F. Analysis of pectin mutants and natural accessions of Arabidopsis highlights the impact of de-methyl-esterified homogalacturonan on tissue saccharification. Biotechnol. Biofuel 2013, 6, 163. [Google Scholar] [CrossRef] [PubMed]




| Lignocellulosic Biomass | Cellulose (%) | Hemicellulose (%) | Lignin (%) |
|---|---|---|---|
| Rice straw | 15–29 | 9–17 | 12–18 |
| Wheat straw | 34–39 | 21–34 | 22–25 |
| Corn stover | 21–37 | 22–31 | 14–20 |
| Switch grass | 31–38 | 26–34 | 18–22 |
| Reed | 34–36 | 26 | 21 |
| Sugarcane bagasse | 42–45 | 25–28 | 20 |
| Miscanthus | 20–40 | 23–35 | 19–31 |
| Rapeseed | 20–35 | 16–20 | 15–24 |
| S. No. | Species | Target Genes | Wall Polymer and Its Regulation | Phenotypic Modifications | Pretreatment | Target for Pretreatment | References | ||
|---|---|---|---|---|---|---|---|---|---|
| Cell Wall Structure and Composition | Biomass Yield | Enzymatic Digestibility | |||||||
| 1 | Arabidopsis | AtCesA1, 3 | Cellulose synthesis | Decrease in cellulose and crystallinity index | / | + | ND | NA | [40] |
| 2 | Tobacco | AtCESA3 | Cellulose synthesis | No significant effect on cell wall composition | - | + | ND | NA | [41] |
| 3 | B. distachyon | BdCesA4, 7 | Cellulose synthesis | Decrease in cellulose and crystallinity index | - | / | ND | NA | [42] |
| 4 | Barley | HvCesA4, 8 | Cellulose synthesis | Decrease in cellulose and crystallinity index | - | NA | ND | NA | [21] |
| 5 | Arabidopsis | PttCel9A1 | Cellulose degradation | Decrease in cellulose and crystallinity index | + | NA | ND | NA | [47] |
| 6 | Arabidopsis | PtGH9B5, C2 | Cellulose degradation | Increase in cellulose | - | NA | ND | NA | [6] |
| 7 | Arabidopsis | AtGH9C2 | Cellulose degradation | Decrease in cellulose and crystallinity index | + | NA | ND | NA | [6] |
| 8 | Rice | OsMYB103L | Cellulose regulation | Increased secondary wall thickness | / | NA | ND | NA | [43] |
| 9 | Rice | OsMYB61; NAC29, 31 | Cellulose regulation | Increased secondary wall thickness | / | NA | ND | NA | [44] |
| 10 | Rice | OsXAX1 (GT61) | Hemicellulose synthesis | Decrease in Xyl, ferulic, coumaric acid | - | + | ND | NA | [45] |
| 11 | Rice | OsIRX10 (GT47) | Hemicellulose synthesis | Reduced ratio of xylose and arabinose, affect growth | - | + | Hot water | Lignin | [7] |
| 12 | Arabidopsis | AtTBL129 | Hemicellulose synthesis | Decrease in acetate content | NA | - | ND | NA | [48] |
| 13 | Arabidopsis | AtESK1(DUF231) | Hemicellulose synthesis | Decrease in acetylated xylan | - | + | ND | NA | [8] |
| 14 | Arabidopsis | AnAXE1 | Hemicellulose synthesis | Decrease in xylan content | / | + | Hot water and alkali | Lignin | [46] |
| 15 | Arabidopsis | AtMUR3/AtMURUS3 (GT47) | Hemicellulose synthesis | Decrease in xyloglucan | - | NA | ND | NA | [49] |
| 16 | Arabidopsis | AtAXY9(TBL) | Hemicellulose synthesis | Decrease in xyloglucan | - | NA | ND | NA | [50] |
| 17 | Poplar | PtGAUT12.1,12.2(GT8) | Hemicellulose synthesis | Reduce xylan content | + | + | Thermal pretreatment at 180 °C | Lignin | [23] |
| 18 | Poplar | AtKNAT7 | Hemicellulose degradation | Increased expression of IRX8, IRX9, FRA8 | - | NA | ND | NA | [51] |
| 19 | Alfalfa | MsHCT | Lignin synthesis | Decreased lignin content; increased H | - | NA | ND | NA | [52] |
| 20 | Arabidopsis | AtCSE-1 | Lignin synthesis | Decreased lignin content and G monomer, increased H | - | + | ND | NA | [53] |
| 21 | B. distachyon | BdCAD1 | Lignin synthesis | Decrease in lignin content and S unit, increase in H and G unit | / | + | ND | NA | [54] |
| 22 | Poplar | PtCCR | Lignin synthesis | Decrease in lignin content, increase in G unit and hemicellulose | - | + | Alkaline and acid | Lignin | [55] |
| 23 | Sugarcane | SbCOMT | Lignin synthesis | Decreased lignin content | - | + | Acid | Lignin | [56] |
| 24 | Switch grass | SbCOMT | Lignin synthesis | Decreased lignin content, S and G ratio, increased hemicellulose | / | + | ND | NA | [57] |
| 25 | Poplar | AsFMT | Lignin interlinking | Increased ferulic acid conjugates, ester bonds and Degradation of β-ether bonds | / | + | Alkaline | Lignin | [37] |
| 26 | Arabidopsis | AtREF4/AtRFR1 | Lignin regulation | Increase in G unit, reduction of S unit and S/G ratio | NA | - | Hot water | Lignin | [58] |
| 27 | Arabidopsis | AtQUA2-1 | Pectin synthesis | Decrease of GalA and de-methyl-esterified HG, increase Gal and Xyl | - | + | ND | NA | [59] |
| 28 | Poplar | PtGAUT12 | Pectin synthesis | Decreased Xyl, GalA, HG, RG; increased Man and Gal | + | + | Thermal pretreatment at 180 °C | Lignin | [23] |
| 29 | Arabidopsis | PcPL1 | Pectin degradation | Increase Glc; affect growth | - | + | ND | NA | [32] |
| S. No. | Plant | Modified Gene | Outcomes | Reference |
|---|---|---|---|---|
| 1 | Arabidopsis | AtCesA1, 3 | 8% decrease in crystallanity | [40] |
| 2 | Tobacco | AtCESA3 | 54%–66% increase in enzymatic saccharification | [41] |
| 3 | Arabidopsis | PtGH9B5 | slight decreases in each carbohydrate | [6] |
| 4 | Rice | OsMYB103L | 13% decrease in cellulose content, | [43] |
| 5 | Rice | OsIRX10 (GT47) | 10% reduction in xylose, 25 and 87% reduction in thickness of primary and secondary cell wall | [7] |
| 6 | Arabidopsis | AtESK1(DUF231) | smaller rosette leaves and shorter inflorescence stems, 35% reduction in secondary wall thickness | [8] |
| 7 | Arabidopsis | AtAXY9(TBL) | 35% decrease in xylan, 18% decrease in pectin fraction | [50] |
| 8 | Sugarcane | SbCOMT | 29% increase in glucose yield without pretreatment and 34% after pretreatment | [56] |
| 9 | Switch grass | SbCOMT | 34% increase in sugar yield and 28 5 increase in ethanol yield from transgenic line | [57] |
| 10 | Poplar | AsFMT | Almost 2 fold increase in saccharification yield after pretreatment as compared to wild type | [37] |
| 11 | Arabidopsis | AtREF4/AtRFR1 | 30% increase in glucose yield, | [58] |
| 12 | Arabidopsis | AtQUA2-1 | 43% increase in saccharification yield after enzymatic hydrolysis | [59] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, M.; Paritosh, K.; Chawade, A.; Pareek, N.; Vivekanand, V. Genetic Engineering of Energy Crops to Reduce Recalcitrance and Enhance Biomass Digestibility. Agriculture 2018, 8, 76. https://doi.org/10.3390/agriculture8060076
Yadav M, Paritosh K, Chawade A, Pareek N, Vivekanand V. Genetic Engineering of Energy Crops to Reduce Recalcitrance and Enhance Biomass Digestibility. Agriculture. 2018; 8(6):76. https://doi.org/10.3390/agriculture8060076
Chicago/Turabian StyleYadav, Monika, Kunwar Paritosh, Aakash Chawade, Nidhi Pareek, and Vivekanand Vivekanand. 2018. "Genetic Engineering of Energy Crops to Reduce Recalcitrance and Enhance Biomass Digestibility" Agriculture 8, no. 6: 76. https://doi.org/10.3390/agriculture8060076





