Identification of Marbling Gene Loci in Commercial Pigs in Canadian Herds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Phenotypic Analysis
2.2. Genotyping
2.3. Genome-Wide Association Study
2.4. Single SNP Association Analysis
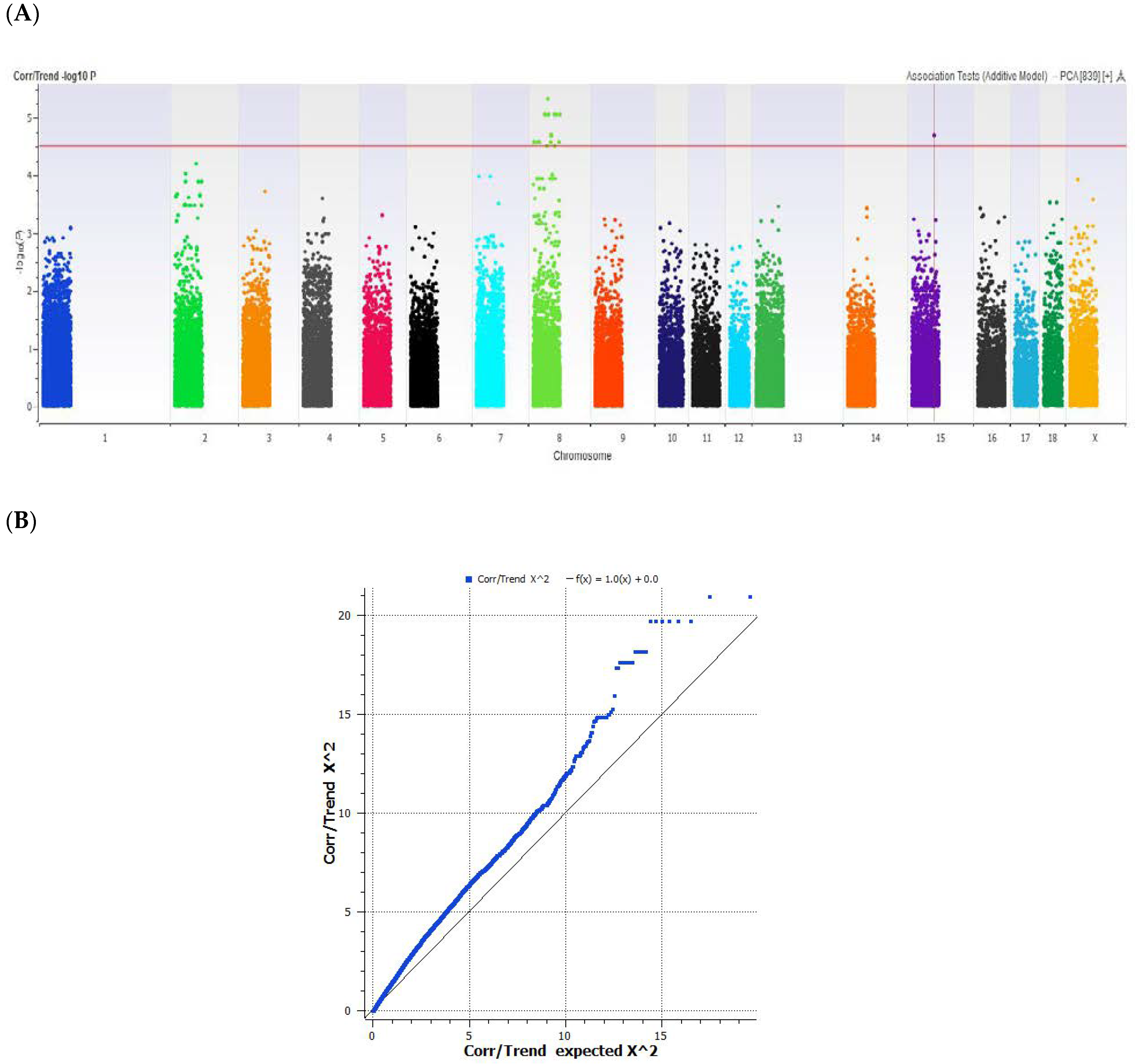
3. Results and Discussion
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Cheng, W.; Cheng, J.; Sun, D.; Pu, H. Marbling analysis for evaluating meat quality: Methods and techniques. Compr. Rev. Food Sci. Food Saf. 2015, 14, 523–535. [Google Scholar] [CrossRef]
- Keeton, J.T.; Hafley, B.S.; Eddy, S.M.; Moser, C.R.; McManus, B.J.; Leffler, T.P. Rapid determination of moisture and fat in meats by microwave and nuclear magnetic resonance analysis—PVM 1:2003. J. AOAC Int. 2003, 86, 1193–1202. [Google Scholar] [PubMed]
- Eikelenboom, G.; Hoving-Bolink, A.H.; Van der Wal, P.G. The eating quality of pork. 2. The influence of intramuscular fat. Fleischwirtschaft 1996, 76, 517–518. [Google Scholar]
- Fortin, A.; Robertson, W.M.; Tong, A.K. The eating quality of Canadian pork and its relationship with intramuscular fat. Meat Sci. 2005, 69, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Cameron, P.J.; Zembayashi, M.; Lunt, D.K.; Mitsuhashi, T.; Mitsumoto, M.; Ozawa, S.; Smith, S.B. Relationship between Japanese beef marbling standard and intramuscular lipid in the m. Longissimus thoracis of Japanese black and American wagyu cattle. Meat Sci. 1994, 38, 361–364. [Google Scholar] [CrossRef]
- USDA. United States standards for grades of pork carcasses. Fed. Regist. Docket 2017, 82, 48971–48976. [Google Scholar]
- Apple, J.K. Nutritional effects on pork quality in swine production. In National Swine Nutrition Guide; Factsheet PIG 12-02-02; U.S. Pork Center of Excellence: Clive, IA, USA, 2015. [Google Scholar]
- Schwab, C.R.; Baas, T.J.; Stalder, K.J. Results from six generations of selection for intramuscular fat in Duroc swine using real-time ultrasound. Ii. Genetic parameters and trends. J. Anim. Sci. 2010, 88, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Solanes, F.X.; Reixach, J.; Tor, M.; Tibau, J.; Estany, J. Genetic correlations and expected response for intramuscular fat content in a Duroc pig line. Livest. Sci. 2009, 123, 63–69. [Google Scholar] [CrossRef]
- Eusebi, P.G.; Gonzalez-Prendes, R.; Quintanilla, R.; Tibau, J.; Cardoso, T.F.; Clop, A.; Amills, M. A genome-wide association analysis for carcass traits in a commercial Duroc pig population. Anim. Genet. 2017, 48, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Sellier, P.; Maignel, L.; Bidanel, J.P. Genetic parameters for tissue and fatty acid composition of backfat, perirenal fat and muscle in large white and landrace pigs. Anim. Int. J. Anim. Biosci. 2010, 4, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Knapp, P.; William, A.; Solkner, J. Genetic parameters for lean meat content and meat quality traits in different pig breeds. Livest. Prod. Sci. 1997, 52, 69–73. [Google Scholar] [CrossRef]
- Liu, X.; Xiong, X.; Yang, J.; Zhou, L.; Yang, B.; Ai, H.; Ma, H.; Xie, X.; Huang, Y.; Fang, S.; et al. Genome-wide association analyses for meat quality traits in Chinese Erhualian pigs and a western Duroc × (landrace × Yorkshire) commercial population. Genet. Sel. Evol. GSE 2015, 47, 44. [Google Scholar] [CrossRef] [PubMed]
- Verardo, L.L.; Sevon-Aimonen, M.L.; Serenius, T.; Hietakangas, V.; Uimari, P. Whole-genome association analysis of pork meat pH revealed three significant regions and several potential genes in Finnish Yorkshire pigs. BMC Genet. 2017, 18, 13. [Google Scholar] [CrossRef] [PubMed]
- Won, S.; Jung, J.; Park, E.; Kim, H. Identification of genes related to intramuscular fat content of pigs using genome-wide association study. Asian Australas. J. Anim. Sci. 2018, 31, 157–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NRC. National Research Council: Nutrient Requirements of Swine; National Academy of Sciences: Washington, DC, USA, 2012; Volume 11, p. 405.
- Canadian Council on Animal Care (CCAC). The CCAC Guideline on: The Care and Use of Farm Animals in Research, Teaching and Testing; CCAC: Ottawa, ON, Canada, 2009; p. 168. [Google Scholar]
- NPPC. National Pork Producers Council: Nutrient Requirements of Swine, 10th ed.; National Academy of Sciences: Washington, DC, USA, 1998. [Google Scholar]
- Juarez, M.; Dugan, M.E.R.; Lopez-Campos, O.; Prieto, N.; Uttaro, B.; Gariepy, C.; Aalhus, J.L. Relative contribution of breed, slaughter weight, sex, and diet to the fatty acid composition of differentiated pork. Can. J. Anim. Sci. 2017, 97, 395–405. [Google Scholar] [CrossRef]
- Faucitano, L.; Rivest, J.; Daigle, J.P.; Levesque, J.; Gariepy, C. Distribution of intramuscular fat content and marbling within the longissimus muscle of pigs. Can. J. Anim. Sci. 2004, 84, 57–61. [Google Scholar] [CrossRef] [Green Version]
- The Pig Genome SusScr11 Assembly. Available online: https://genome.ucsc.edu/ (accessed on 20 February 2018).
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genet. Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- The Sus Scrofa 11.1/SusScr11 Map. Available online: https://genome.ucsc.edu/ (accessed on 20 November 2017).
- NCBI Data Banks. Available online: https://www.ncbi.nlm.nih.gov (accessed on 20 January 2018).
- Brewer, M.S.; Zhu, L.G.; McKeith, F.K. Marbling effects on quality characteristics of pork loin chops: Consumer purchase intent, visual and sensory characteristics. Meat Sci. 2001, 59, 153–163. [Google Scholar] [CrossRef]
- O’Brien, K.F.; Engle, E.C.; Kunkel, L.M. Analysis of human sarcospan as a candidate gene for CFEOM1. BMC Genet. 2001, 2, 3. [Google Scholar]
- Takeda, K.; Takemasa, T. Expression of ammonia transporters RHBG and RHCG in mouse skeletal muscle and the effect of 6-week training on these proteins. Physiol. Rep. 2015, 3, e12596. [Google Scholar] [CrossRef] [PubMed]
- Bertini, E.; D’Amico, A.; Gualandi, F.; Petrini, S. Congenital muscular dystrophies: A brief review. Semin. Pediatr. Neurol. 2011, 18, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Rahimov, F.; Kunkel, L.M. The cell biology of disease: Cellular and molecular mechanisms underlying muscular dystrophy. J. Cell Biol. 2013, 201, 499–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, V.M.; Bode, A.; Chung, S.K.; Gill, J.L.; Nielsen, M.; Cowan, F.M.; Vujic, M.; Thomas, R.H.; Rees, M.I.; Harvey, K.; et al. Novel missense mutations in the glycine receptor beta subunit gene (GLRB) in startle disease. Neurobiol. Dis. 2013, 52, 137–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bittel, D.C.; Kibiryeva, N.; McNulty, S.G.; Driscoll, D.J.; Butler, M.G.; White, R.A. Whole genome microarray analysis of gene expression in an imprinting center deletion mouse model of prader-willi syndrome. Am. J. Med. Genet. 2007, 143A, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Pierce, K.D.; Handford, C.A.; Morris, R.; Vafa, B.; Dennis, J.A.; Healy, P.J.; Schofield, P.R. A nonsense mutation in the alpha1 subunit of the inhibitory glycine receptor associated with bovine myoclonus. Mol. Cell. Neurosci. 2001, 17, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Pattnaik, B.R.; Asuma, M.P.; Spott, R.; Pillers, D.A. Genetic defects in the hotspot of inwardly rectifying k(+) (kir) channels and their metabolic consequences: A review. Mol. Genet. Metab. 2012, 105, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Chen, J.; Cui, J.; Wang, X.; Zhang, X. Polymorphisms on ssc15q21–q26 containing QTL for reproduction in swine and its association with litter size. Genet. Mol. Biol. 2009, 32, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Mikawa, S.; Yoshioka, G. Development of Marbling Pork with Marker-Assisted Selection. Available online: http://www.angrin.tlri.gov.tw/%5C/English/2014Swine/p18-27.pdf (accessed on 20 January 2018).
- Sato, S.; Uemoto, Y.; Kikuchi, T.; Egawa, S.; Kohira, K.; Saito, T.; Sakuma, H.; Miyashita, S.; Arata, S.; Kojima, T.; et al. Snp- and haplotype-based genome-wide association studies for growth, carcass, and meat quality traits in a duroc multigenerational population. BMC Genet. 2016, 17, 60. [Google Scholar] [CrossRef] [PubMed]
- Brenig, B.; Brem, G. Genomic organization and analysis of the 5’ end of the porcine ryanodine receptor gene (RYR1). FEBS Lett. 1992, 298, 277–279. [Google Scholar] [CrossRef]
- Meadus, W.J.; MacInnis, R.; Dugan, M.E.R.; Aalhus, J.L. A PCR-RFLP method to identify the gene in retailed pork chops. Can. J. Anim. Sci. 2002, 82, 449–451. [Google Scholar] [CrossRef]
- Hausman, G.J.; Basu, U.; Du, M.; Fernyhough-Culver, M.; Dodson, M.V. Intermuscular and intramuscular adipose tissues: Bad vs. good adipose tissues. Adipocyte 2014, 3, 242–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puig-Oliveras, A.; Revilla, M.; Castello, A.; Fernandez, A.I.; Folch, J.M.; Ballester, M. Expression-based GWAS identifies variants, gene interactions and key regulators affecting intramuscular fatty acid content and composition in porcine meat. Sci. Rep. 2016, 6, 31803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Developmental progress and current status of the animal QTLDB. Nucleic Acids Res. 2016, 44, D827–D833. [Google Scholar] [CrossRef] [PubMed]
- This Pig Quantitative Trait Locus (QTL) Database. Available online: https://www.animalgenome.org/cgi-bin/QTLdb/SS/index (accessed on 20 January 2018).


| Chromosome | Position | Marker | Base | Nearest Gene | Gene | refSNP | SNP |
|---|---|---|---|---|---|---|---|
| 5 | 47606484 | WU_10.2_5_64313075 | A/G | AK349478 | SSPN | rs322211582 | 8 |
| 7 | 16758370 | ALGA0044073 | A/G | XR_002346095 | ncRNA | n/a | n/a |
| 7 | 43707458 | ASGA0035629 | T/C | XM_003128440 | RhGlycoprotein | rs698040624 | 25 |
| 16 | 18742425 | ALGA0116333 | T/C | AK343791 | Alveolar macrophage | n/a | n/a |
| 16 | 23611505 | ALGA0092214 | T/C | AK396237 | EGFLAM | rs690289509 | 54 |
| 16 | 73722502 | ASGA0097252 | A/G | AK240470 | Uterus | n/a | n/a |
| Position | Marker | Base | Nearest Gene | Gene | refSNP | SNP | |
|---|---|---|---|---|---|---|---|
| 8 | 46622432 | H3GA0025835 | A/G | AJ715855 | GLRB | n/a | n/a |
| 8 | 46677501 | MARC0004983 | T/C | AK396463 | FAM198B | rs705641356 | 24 |
| 15 | 62778504 | CASI0009038 | A/C | AF540391 | KCNJ3 | rs711844269 | 6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meadus, W.J.; Duff, P.; Juarez, M.; Roberts, J.C.; Zantinge, J.L. Identification of Marbling Gene Loci in Commercial Pigs in Canadian Herds. Agriculture 2018, 8, 122. https://doi.org/10.3390/agriculture8080122
Meadus WJ, Duff P, Juarez M, Roberts JC, Zantinge JL. Identification of Marbling Gene Loci in Commercial Pigs in Canadian Herds. Agriculture. 2018; 8(8):122. https://doi.org/10.3390/agriculture8080122
Chicago/Turabian StyleMeadus, William Jon, Pascale Duff, Manuel Juarez, Jordan C. Roberts, and Jennifer L. Zantinge. 2018. "Identification of Marbling Gene Loci in Commercial Pigs in Canadian Herds" Agriculture 8, no. 8: 122. https://doi.org/10.3390/agriculture8080122





