Deep Transfer Learning for Modality Classification of Medical Images
Abstract
:1. Introduction
2. Related Work
3. Methods
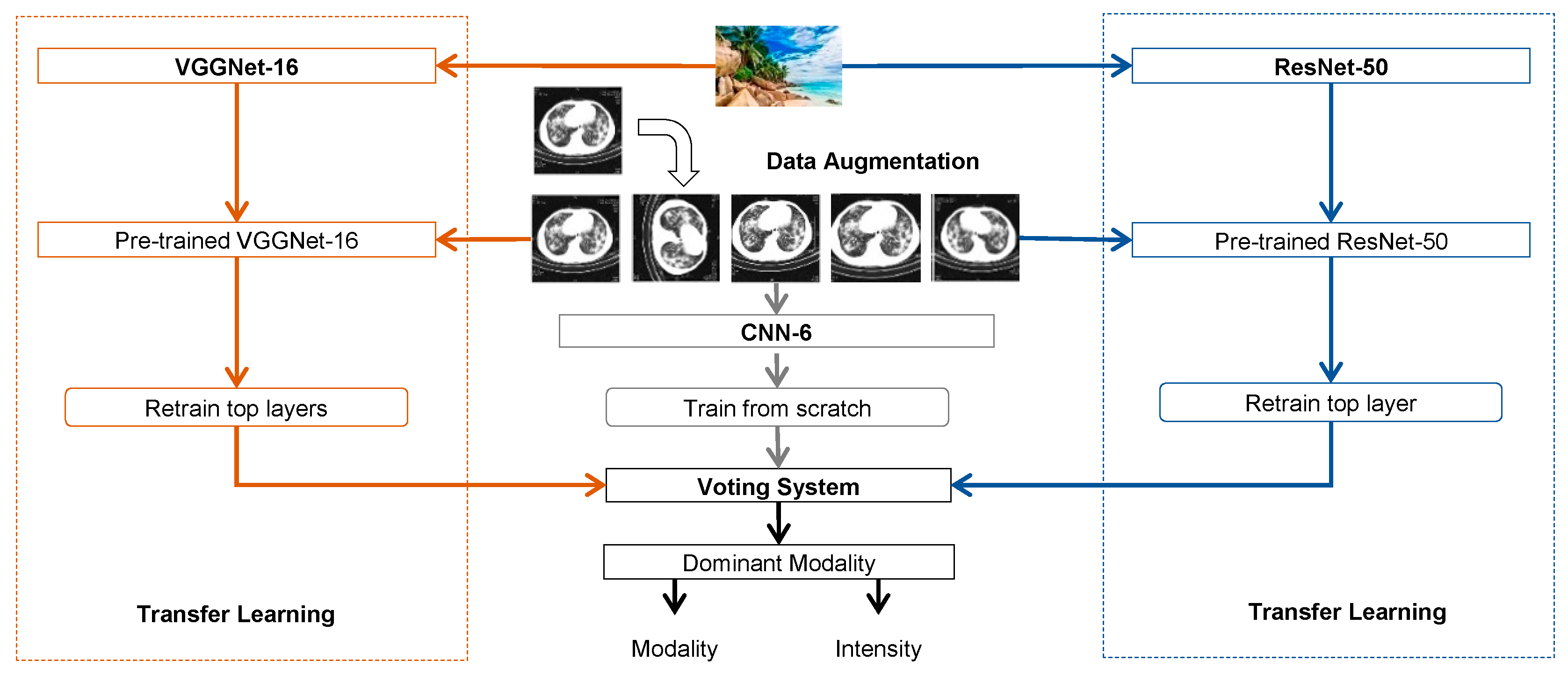
3.1. Convolutional Neural Networks
3.2. Transfer Learning
3.3. Data Augmentation
3.4. Voting System
4. Experiments
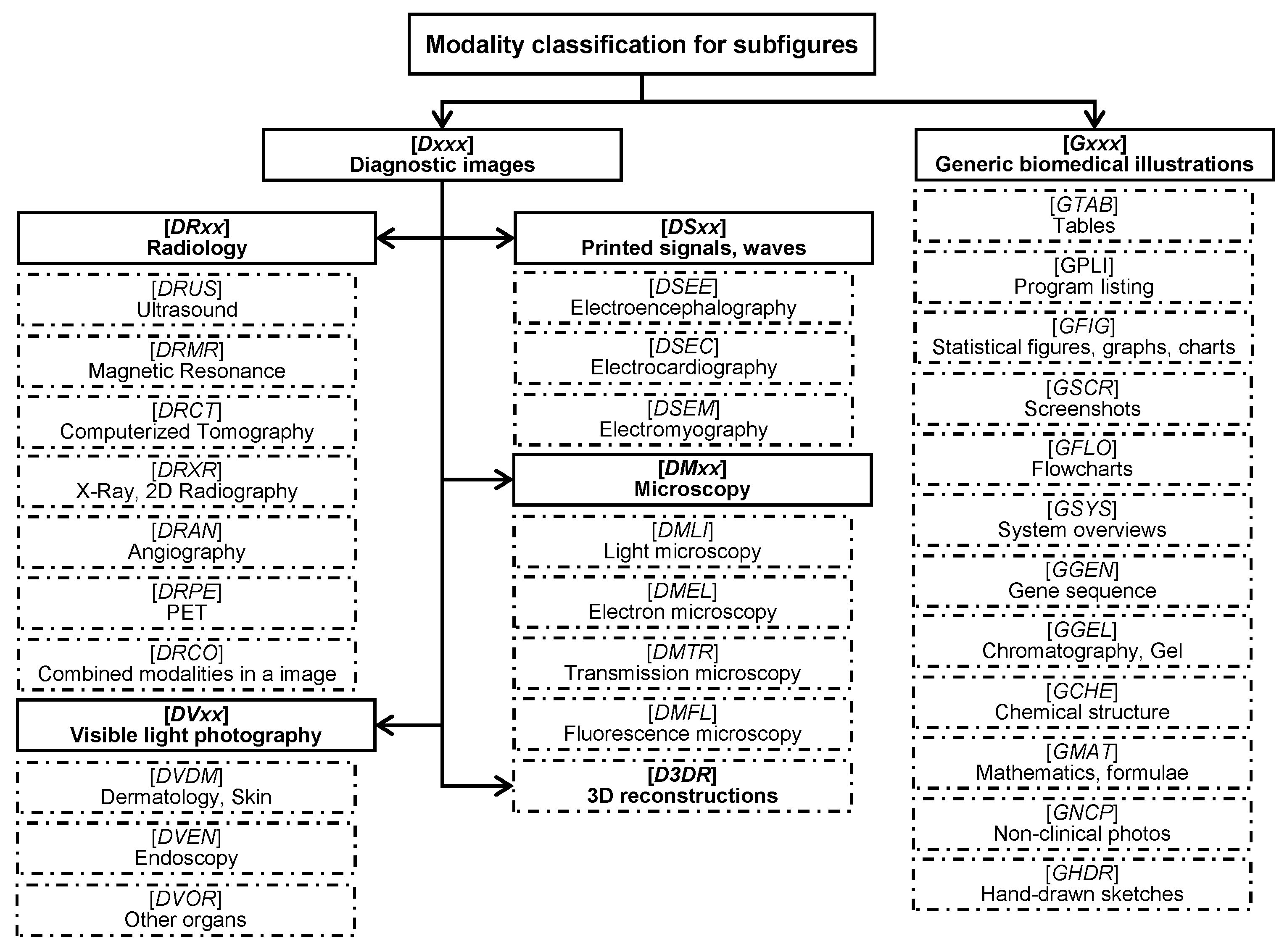
4.1. Datasets
4.2. Baselines
4.3. Expeimental Setup
4.4. Experimental Results and Discussion
4.4.1. Deep Transfer Learning
4.4.2. Data Augmentation
4.4.3. Running Time
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lu, Z. PubMed and beyond: A survey of web tools for searching biomedical literature. Database 2011. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.F.; Saeed, A.; Haider, S.; Ahmed, K.; Ahmed, A. Application of medical images for diagnosis of diseases-review article. World J. Microbiol. Biotechnol. 2017, 2, 135–138. [Google Scholar]
- Shi, J.; Zheng, X.; Li, Y.; Zhang, Q.; Ying, S. Multimodal Neuroimaging Feature Learning with Multimodal Stacked Deep Polynomial Networks for Diagnosis of Alzheimer’s Disease. IEEE J. Biomed. Health Inform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wu, J.; Li, Y.; Zhang, Q.; Ying, S. Histopathological image classification with color pattern random binary hashing based PCANet and matrix-form classifier. IEEE J. Biomed. Health Inform. 2016. [Google Scholar] [CrossRef] [PubMed]
- De Herrera, A.G.S.; Kalpathy-Cramer, J.; Fushman, D.D.; Antani, S.; Müller, H. Overview of the ImageCLEF 2013 medical tasks. In Proceedings of the Working Notes of CLEF, Valencia, Spain, 23–26 September 2013. [Google Scholar]
- Müller, H.; Michoux, N.; Bandon, D.; Geissbuhler, A. A review of content-based image retrieval systems in medical applications—Clinical benefits and future directions. Int. J. Med. Inform. 2004, 73, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Demner-Fushman, D.; Antani, S.; Simpson, M.; Thoma, G.R. Design and development of a multimodal biomedical information retrieval system. J. Comput. Sci. Eng. 2012, 6, 168–177. [Google Scholar] [CrossRef]
- Tirilly, P.; Lu, K.; Mu, X.; Zhao, T.; Cao, Y. On modality classification and its use in text-based image retrieval in medical databases. In Proceedings of the 9th International Workshop on Content-Based Multimedia Indexing (CBMI) 2011, Madrid, Spain, 13–15 June 2011. [Google Scholar]
- De Herrera, A.G.S.; Markonis, D.; Müller, H. Bag-of-Colors for Biomedical Document Image Classification. In Medical Content-Based Retrieval for Clinical Decision Support (MCBR-CDS) 2012, Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Pelka, O.; Friedrich, C.M. FHDO biomedical computer science group at medical classification task of ImageCLEF 2015. In Proceedings of the Working Notes of CLEF, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Cirujeda, P.; Binefa, X. Medical Image Classification via 2D color feature based Covariance Descriptors. In Proceedings of the Working Notes of CLEF, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Valavanis, L.; Stathopoulos, S.; Kalamboukis, T. IPL at CLEF 2016 Medical Task. In Proceedings of the Working Notes of CLEF, Évora, Portugal, 5–8 September 2016. [Google Scholar]
- Li, P.; Sorensen, S.; Kolagunda, A.; Jiang, X.; Wang, X.; Kambhamettu, C.; Shatkay, H. UDEL CIS Working Notes in ImageCLEF 2016. In Proceedings of the Working Notes of CLEF, Évora, Portugal, 5–8 September 2016. [Google Scholar]
- Pelka, O.; Friedrich, C.M. Modality prediction of biomedical literature images using multimodal feature representation. GMS Med. Inform. Biom. Epidemiol. 2016, 12. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. Comput. Sci. 2014; arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Hynes Convention Center, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 27–30 June 2016. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; Berg, A.C.; Li, F.F. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Lin, H.; Meng, J.; Zhao, Z.; Li, Y.; Zuo, L. Modality classification for medical images using multiple deep convolutional neural networks. J. Colloid Interface Sci. 2015, 11, 5403–5413. [Google Scholar] [CrossRef]
- Ravishankar, H.; Sudhakar, P.; Venkataramani, R.; Thiruvenkadam, S.; Annangi, P.; Babu, N.; Vaidya, V. Understanding the Mechanisms of Deep Transfer Learning for Medical Images. In Deep Learning and Data Labeling for Medical Applications. Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2016; pp. 188–196. [Google Scholar]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kim, J.; Lyndon, D.; Fulham, M.; Feng, D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J. Biomed. Health Inform. 2017, 21, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xia, Y.; Wu, Q.; Xie, Y. Classification of Medical Images and Illustrations in the Biomedical Literature Using Synergic Deep Learning. Comput. Sci. 2017; arXiv:1706.09092. [Google Scholar]
- Koitka, S.; Friedrich, C.M. Traditional feature engineering and deep learning approaches at medical classification task of ImageCLEF 2016. In Proceedings of the Working Notes of CLEF, Évora, Portugal, 5–8 September 2016. [Google Scholar]
- Donahue, J.; Jia, Y.; Vinyals, O.; Hoffman, J.; Zhang, N.; Tzeng, E.; Darrell, T. Decaf: A deep convolutional activation feature for generic visual recognition. In Proceedings of the International Conference on Machine Learning, Beijing, China, 21–26 June 2014. [Google Scholar]
- Lowe, D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Wengert, C.; Douze, M.; Jégou, H. Bag-of-colors for improved image search. In Proceedings of the 19th ACM International Conference on Multimedia, New York, NY, USA, 28 November–1 December 2011. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Jiang, Y.G.; Hauptmann, A.G.; Ngo, C.W. Evaluating bag-of-visual-words representations in scene classification. In Proceedings of the International Workshop on ACM Multimedia Information Retrieval, University of Augsburg, Augsburg, Germany, 28–29 September 2007. [Google Scholar]
- Yin, X.; Düntsch, I.; Gediga, G. Quadtree representation and compression of spatial data. Trans. Rough Sets XIII 2011, 207–239. [Google Scholar] [CrossRef]
- De Herrera, A.G.S.; Müller, H.; Bromuri, S. Overview of the ImageCLEF 2015 medical classification task. In Proceedings of the Working Notes of CLEF, Toulouse, France, 8–11 September 2015. [Google Scholar]
- De Herrera, A.G.S.; Schaer, R.; Bromuri, S.; Müller, H. Overview of the ImageCLEF 2016 medical task. In Proceedings of the Working Notes of CLEF, Évora, Portugal, 5–8 September 2016. [Google Scholar]
- Yu, Y.; Lin, H.; Meng, J.; Zhao, Z. Visual and Textual Sentiment Analysis of a Microblog Using Deep Convolutional Neural Networks. Algorithms 2016, 9, 41. [Google Scholar] [CrossRef]
- Yu, Y.; Lin, H.; Meng, J.; Wei, X.; Zhao, Z. Assembling Deep Neural Networks for Medical Compound Figure Detection. Information 2017, 8, 48. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the 13th International Conference on Artificial Intelligence and Statistics (AISTATS), Sardinia, Italy, 13–15 May 2010. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. Comput. Sci. 2014; arXiv:1412.6980. [Google Scholar]
- Kuncheva, L.I.; Rodríguez, J.J. A weighted voting framework for classifiers ensembles. Knowl. Inf. Syst. 2014, 38, 259. [Google Scholar] [CrossRef]
- Chen, D.; Riddle, D.L. Function of the PHA-4/FOXA transcription factor during C. elegans post-embryonic development. BMC Dev. Biol. 2008, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.; Kalpathy-Cramer, J.; Demner-Fushman, D.; Antani, S. Creating a classification of image types in the medical literature for visual categorization. In Proceedings of the SPIE Medical Imaging, San Francisco, CA, USA, 21–26 January 2012. [Google Scholar]
| Methods | 10FCV | Evaluation | ||
|---|---|---|---|---|
| DS_Original | DS_Original | DS_Aug1 | DS_Aug2 | |
| Baseline_2015 [10,14] | - | - | 60.91 | - |
| CNN-6 | 59.95 | 57.09 | 58.33 | 66.13 |
| VGGNet-16 | 87.27 | 67.83 | 70.50 | 71.61 |
| ResNet-50 | 89.34 | 72.10 | 75.75 | 76.78 |
| Our proposed model | 90.22 | 72.42 | 76.07 | 76.87 |
| Methods | 10FCV | Evaluation | ||
|---|---|---|---|---|
| DS_Original | DS_Original | DS_Aug1 | DS_Aug2 | |
| Baseline_2016 [23] | - | - | 85.38 | - |
| CNN-6 | 75.87 | 70.67 | 74.70 | 81.86 |
| VGGNet-16 | 85.13 | 78.99 | 81.73 | 83.54 |
| ResNet-50 | 87.47 | 82.51 | 85.25 | 86.92 |
| Our proposed model | 88.40 | 82.61 | 86.07 | 87.37 |
| CNNs | Epochs | Learning Rate | Batch Size | |||
|---|---|---|---|---|---|---|
| 10FCV | DS_Original | DS_Aug1 | DS_Aug2 | |||
| CNN-6 | 25 | 25 | 25 | 5 | 0.001 | 32 |
| VGGNet-16 | 15 | 15 | 15 | 5 | 0.0002 | 32 |
| ResNet-50 | 30 | 30 | 30 | 5 | 0.0002 | 32 |
| Methods | Training (ms) | Test (ms) |
|---|---|---|
| CNN-6 | 18.2 | 4 |
| VGGNet-16 | 28.4 | 20.5 |
| ResNet-50 | 29 | 10.7 |
| Methods | ImageCLEF2015 (s) | ImageCLEF2016 (s) | ||
|---|---|---|---|---|
| DS_Aug1 | DS_Aug2 | DS_Aug1 | DS_Aug2 | |
| CNN-6 | 143 | 4315 | 170 | 4688 |
| VGGNet-16 | 224 | 5757 | 280 | 6299 |
| ResNet-50 | 229 | 6532 | 267 | 7112 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Lin, H.; Meng, J.; Wei, X.; Guo, H.; Zhao, Z. Deep Transfer Learning for Modality Classification of Medical Images. Information 2017, 8, 91. https://doi.org/10.3390/info8030091
Yu Y, Lin H, Meng J, Wei X, Guo H, Zhao Z. Deep Transfer Learning for Modality Classification of Medical Images. Information. 2017; 8(3):91. https://doi.org/10.3390/info8030091
Chicago/Turabian StyleYu, Yuhai, Hongfei Lin, Jiana Meng, Xiaocong Wei, Hai Guo, and Zhehuan Zhao. 2017. "Deep Transfer Learning for Modality Classification of Medical Images" Information 8, no. 3: 91. https://doi.org/10.3390/info8030091





