Anomalous Diffusion within the Transcriptome as a Bio-Inspired Computing Framework for Resilience
Abstract
:1. Introduction
2. Materials and Methods
3. Results
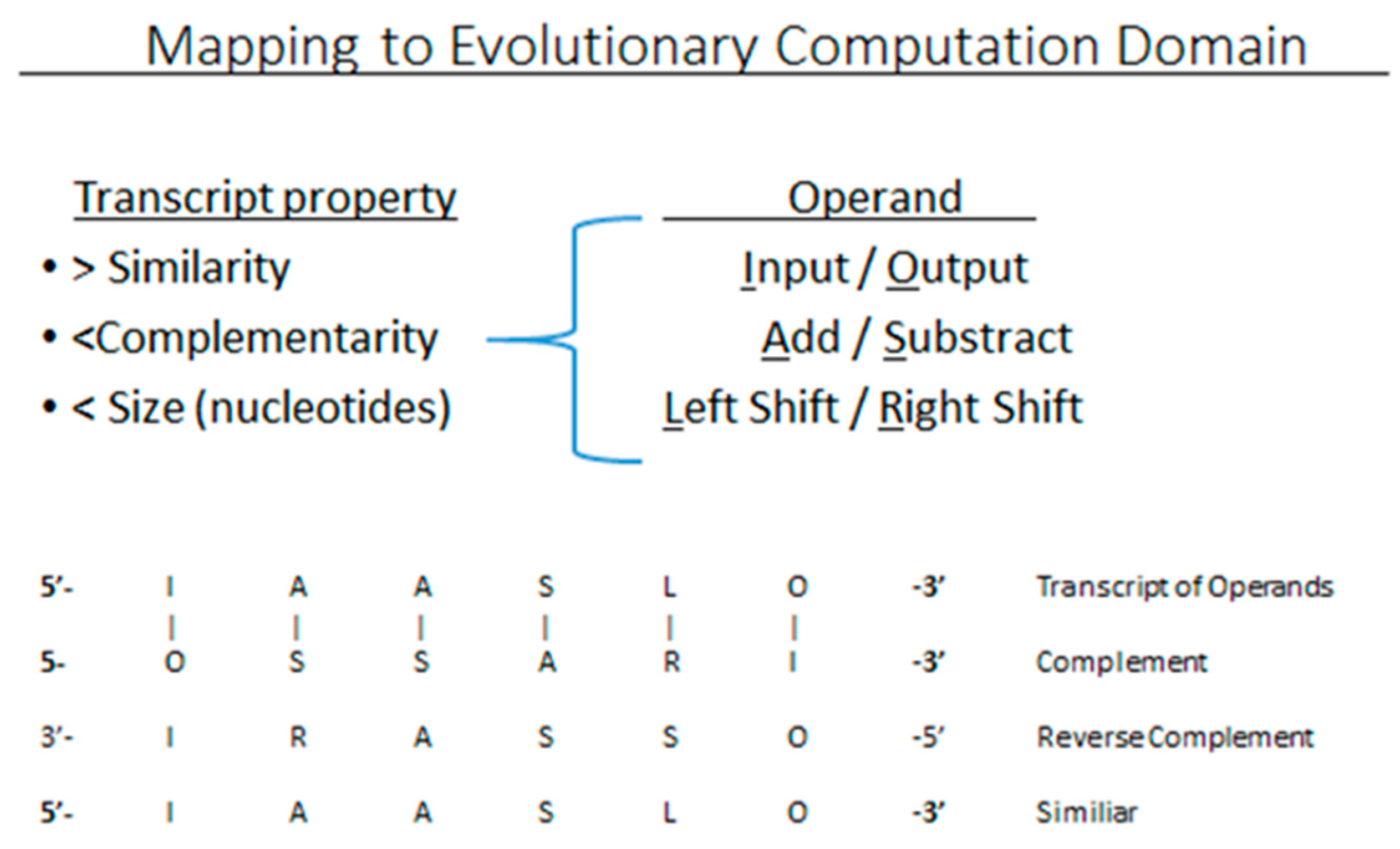
3.1. Evolutionary Computing
3.2. Dynamic Optimization Strategies
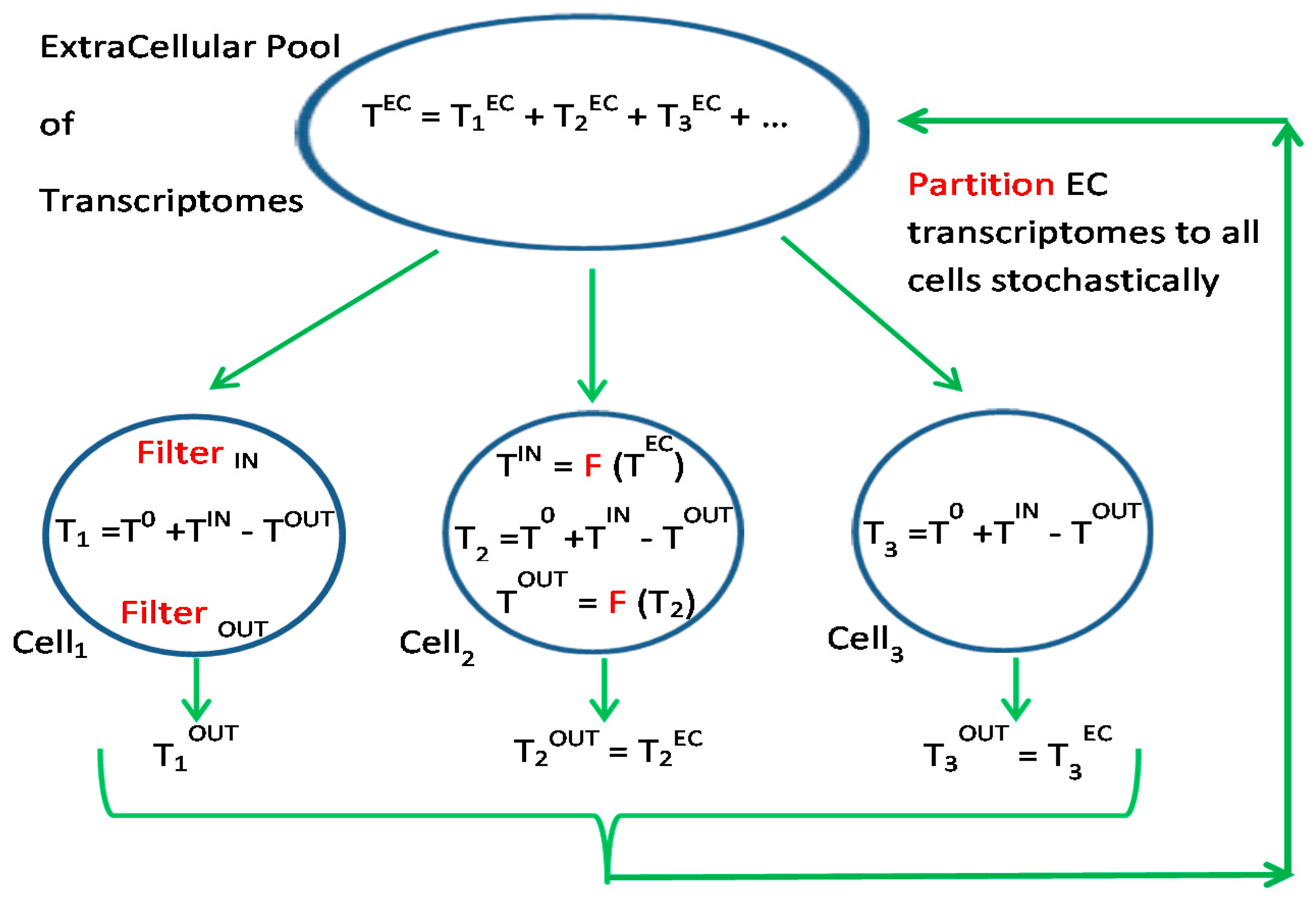
3.3. Silencing and Enhancing Transcripts for Dynamic Environments
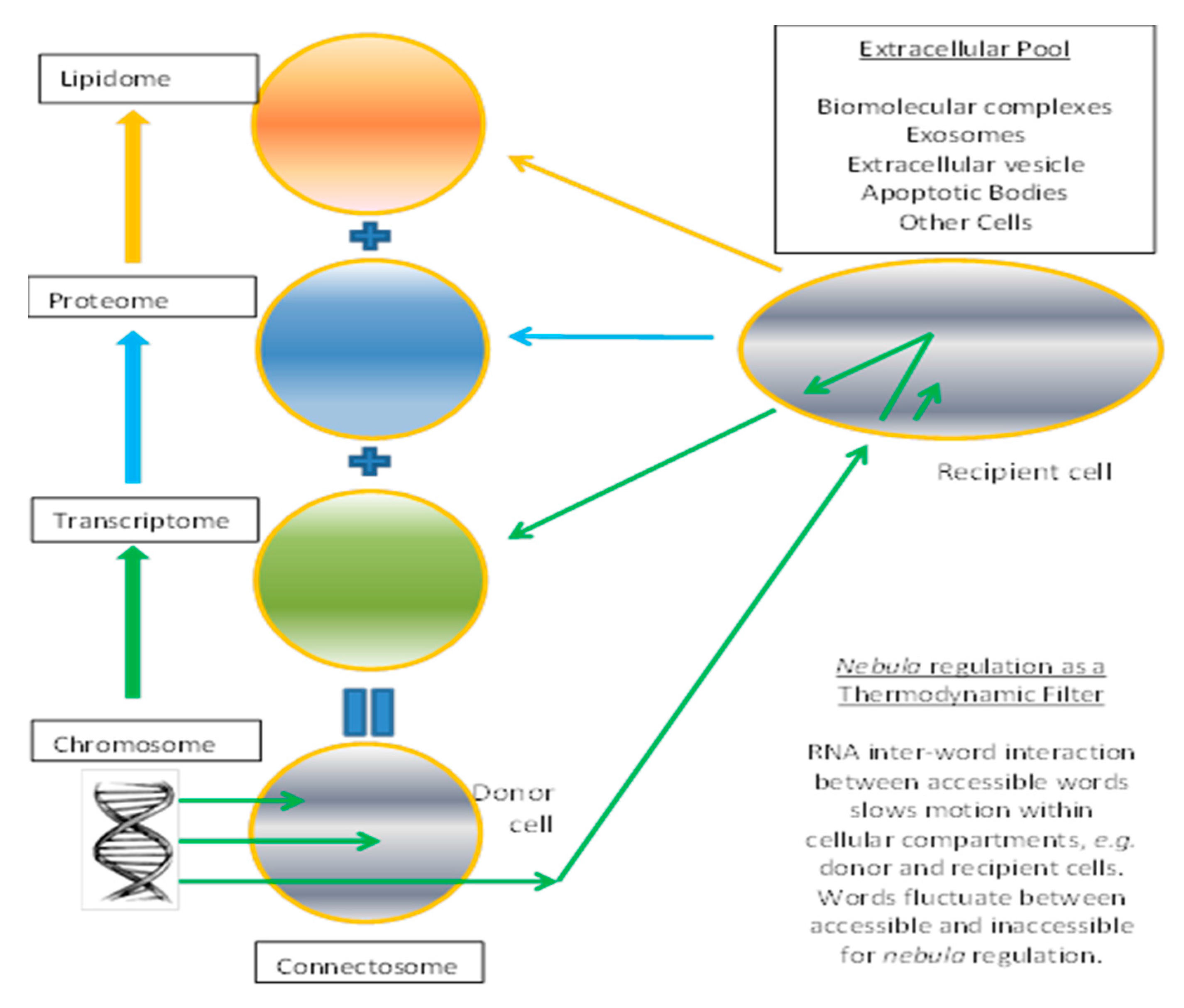
3.4. Connectosome in the Grand Ensemble
3.5. Relevance to Membrane Computing
3.6. In Silico Virtual Living System as a Persistent Turning Machine (PTM)
3.7. Anomalous Diffusion Model
4. Discussion
4.1. Resilience as a Systems Biology Measure from Transcriptome Model
4.2. Measuring Complexity
4.3. Measuring Resilience
5. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Talbi, E.-G. Metaheuristics: From Design to Implementation; John Wiley & Sons: Hoboken, NJ, USA, 2009; Volume 74. [Google Scholar]
- Osman, I.H.; Laporte, G. Metaheuristics: A bibliography. Ann. Oper. Res. 1996, 63, 513–623. [Google Scholar]
- Holland, J.H. Adaptation in Natural and Artificial Systems; University of Michigan Press: Ann Arbor, MI, USA, 1975. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle Swarm Optimization. In Proceedings of the IEEE International Conference on Neural Networks, Perth, Australia, 27 November–1 December 1995; pp. 1942–1948. [Google Scholar]
- Dorigo, M.; Stützle, T. Ant Colony Optimization; Bradford Company: Holland, MI, USA, 2004. [Google Scholar]
- Boussaïd, I.; Lepagnot, J.; Siarry, P. A survey on optimization metaheuristics. Inf. Sci. 2013, 237, 82–117. [Google Scholar] [CrossRef]
- Ishibuchi, H.; Yoshida, T.; Murata, T. Balance between genetic search and local search in memetic algorithms for multiobjective permutation flowshop scheduling. IEEE Trans. Evol. Comput. 2003, 7, 204–223. [Google Scholar] [CrossRef]
- Wolpert, D.H.; Macready, W.G. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef]
- Wang, X.-Q.; Abebe, F.; Seffens, W. Dynamic System Modeling the Whole Transcriptome in a Eukaryotic Cell. In Proceedings of the Dynamic Systems and Applications, Atlanta, GA, USA, 27–30 May 2015; Volume 7, pp. 342–344. [Google Scholar]
- Seffens, W. Models of RNA Interaction from Experimental Datasets: Framework of Resilience; Marchi, F.A., Cirillo, P.D.R., Mateo, E.C.C., Eds.; InTech Publishing: Rijeka, Croatia, 2017; Chapter in Transcriptome Analysis; ISBN 978-953-51-5452-5. [Google Scholar]
- Batagov, A.; Kuznetsov, V.; Kurochkin, I. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genom. 2011, 12 (Suppl. 3), S18. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Kim, O.; Gho, Y. Extracellular vesicles as emerging intercellular communicasomes. BMB Rep. 2014, 47, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Shifrin, D.; Beckler, M.; Coffey, R.; Tyska, M. Extracellular vesicles: Communication, coercion, and conditioning. Mol. Biol. Cell 2013, 24, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Regner, B.; Vucinic, D.; Domnisoru, C.; Bartol, T.; Hetzer, M.; Tartakovsky, D.; Sejnowski, T. Anomalous diffusion of single particles in cytoplasm. Biophys. J. 2013, 104, 1652–1660. [Google Scholar] [CrossRef] [PubMed]
- Seffens, W.; Abebe, F.; Evans, C.; Wang, X.-Q. Spatial Partitioning of miRNAs is related to Sequence Similarity in Overall Transcriptome. Int. J. Mol. Sci. 2016, 17, 830. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Park, C.W.; Zeng, Y.; Zhang, X.; Subramanian, S.; Steer, C. Mature microRNAs identified in highly purified nuclei from HCT116 colon cancer cells. RNA Biol. 2010, 7, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- Eiben, A.; Smith, J. Introduction to Evolutionary Computing; Springer: Berlin, Germany, 2003. [Google Scholar]
- Banzhaf, W.; Beslon, G.; Christensen, S.; Foster, J.; Képès, F.; Lefort, V.; Miller, J.; Radman, M.; Ramsden, J. From Artificial Evolution to Computational Evolution: A Research Agenda. Nat. Rev. Genet. 2006, 7, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Timmis, J.; Amos, M.; Banzhaf, W.; Tyrrell, A. Going back to our Roots: Second Generation Biocomputing. Int. J. Unconv. Comput. 2006, 2, 349–382. [Google Scholar]
- Cruz, C.; González, J.R.; Pelta, D. Optimization in dynamic environments: A survey on problems, methods and measures. Soft Comput. 2011, 15, 1427–1448. [Google Scholar] [CrossRef]
- Pelta, D.; Cruz, C.; Verdegay, J. Simple control rules in a cooperative system for dynamic optimization problems. Int. J. Gen. Syst. 2009, 38, 701–717. [Google Scholar] [CrossRef]
- Novoa-Hernández, P.; Corona, C.; Pelta, D. Efficient multi-swarm PSO algorithms for dynamic environments. Memet. Comput. 2011, 3, 163–174. [Google Scholar] [CrossRef]
- Jin, Y.; Branke, J. Evolutionary optimization in uncertain environments—A survey. IEEE Trans. Evol. Comput. 2005, 9, 303–317. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Yang, S.; Branke, J. Evolutionary dynamic optimization: A survey of the state of the art. Swarm Evol. Comput. 2012, 6, 1–24. [Google Scholar] [CrossRef]
- Angeline, P. Adaptive and Self-Adaptive Evolutionary Computations. In Computational Intelligence: A Dynamic Systems Perspective; Palaniswami, M., Attikiouzel, Y., Eds.; IEEE Press: Hoboken, NJ, USA, 1995; pp. 152–163. [Google Scholar]
- Eiben, A.; Michalewicz, Z.; Schoenauer, M.; Smith, J. Parameter Control in Evolutionary Algorithms. In Parameter Setting in Evolutionary Algorithms, Studies in Computational Intelligence; Lobo, F., Lima, C., Michalewicz, Z., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 54, pp. 47–75. [Google Scholar]
- Meyer-Nieberg, S.; Beyer, H.-G. Self-Adaptation in Evolutionary Algorithms. In Parameter Setting in Evolutionary Algorithms, Studies in Computational Intelligence; Lobo, F., Lima, C., Michalewicz, Z., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 54, pp. 19–46. [Google Scholar]
- Weicker, K.; Weicker, N. On Evolution Strategy Optimization in Dynamic Environments. In Proceedings of the 1999 Congress on Evolutionary Computation, Washington, DC, USA, 6–9 July 1999; IEEE Press: Hoboken, NJ, USA; pp. 2039–2046. [Google Scholar]
- Miller, J.F. Cartesian Genetic Programming (Natural Computing Series); Springer: Berlin, Germany, 2011. [Google Scholar]
- Singh, Y.H.; Andrabi, M.; Kahali, B.; Ghosh, C.; Mizuguchi, K.; Kochetov, A.; Ahmad, S. On nucleotide solvent accessibility in RNA structure. Gene 2010, 463, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Seffens, W.; Digby, D. mRNAs Have Greater Calculated Folding Free Energies than Shuffled or Codon Choice Randomized Sequences. Nucleic Acids Res. 1999, 27, 1578–1584. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.-K.; Digby, D.; Davis, A.; Seffens, W. Whole Transcriptome mRNA Secondary Structure Analysis Using Distributed Computation. In Proceedings of the 2006 IEEE International Conference on Granular Computing, Atlanta, GA, USA, 10–12 May 2006; pp. 647–650. [Google Scholar]
- Kaufmann, P.; Platzner, M. Multi-Objective Intrinsic Evolution of Embedded Systems. In Organic Computing—A Paradigm Shift for Complex Systems; Muller-Schloer, C., Schmeck, H., Ungerer, T., Eds.; Springer Basel AG: Basel, Switzerland, 2011; pp. 193–206. [Google Scholar]
- Kaufmann, P.; Platzner, M. Advanced Techniques for the Creation and Propagation of Modules in Cartesian Genetic Programming. In Genetic and Evolutionary Computation (GECCO); ACM: New York, NY, USA, 2008; pp. 1219–1226. [Google Scholar]
- Paun, G.H. Computing with Membranes; Technical Report; Turku Center for Computer Science: Turku, Finland, 1998. [Google Scholar]
- Nishida, T.Y. An approximate algorithm for NP-complete optimization problems exploiting P systems. In Proceedings of the Brainstorming Workshop on Uncertainty in Membrane Computing, Palma, Majorca, Spain, 8–10 November 2004; pp. 185–192. [Google Scholar]
- Garcia-Arnau, M.; Manrique, D.; Rodriguez-Paton, A.; Sosík, P. A P system and a constructive membrane-inspired DNA algorithm for solving the maximum clique problem. BioSystems 2007, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.Q.; Alhazov, A. Solving HPP and SAT by P systems with active membrane and separation rules. Acta Inform. 2006, 43, 131–145. [Google Scholar] [CrossRef]
- Niu, Y.Y.; Pan, L.Q.; Perez-Jimenez, M.J.; Font, M.R. A Tissue P Systems Based Uniform Solution to Tripartite Matching Problem. Fundam. Inform. 2011, 109, 1–10. [Google Scholar]
- Haddow, P.C.; Tyrrell, A.M. Genetic Programming and Evolvable Machines; Springer: Berlin, Germany, 2011; Volume 12, pp. 183–215. [Google Scholar]
- Goldin, D.; Smolka, S.; Wegner, P. Turing Machines, transition systems, and interaction. Electr. Notes Theor. Comput. Sci. 2001, 52, 120–136. [Google Scholar] [CrossRef]
- Turner, A.J.; Miller, J.F. Recurrent Cartesian Genetic Programming. In Parallel Problem Solving from Nature—PPSN XIII 2014 LNCS 8672; Bartz-Beielstein, T., Branke, J., Filipič, B., Smith, J., Eds.; Springer International Publishing AG: Cham, Switzerland, 2014; pp. 476–486. [Google Scholar]
- Banzhaf, W. Genetic Programming and Emergence. In Genetic Programming and Evolvable Machines; Springer International Publishing AG: Cham, Switzerland, 2013. [Google Scholar]
- Trovato, F.; Tozzini, V. Diffusion within the cytoplasm: A mesoscale model of interacting macromolecules. Biophys. J. 2014, 107, 2579–2591. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ari, Y.; Brody, Y.; Kinor, N.; Mor, A.; Tsukamoto, T.; Spector, D.L.; Singer, R.H.; Shav-Tal, Y. The life of an mRNA in space and time. J. Cell Sci. 2010, 123, 1761–1774. [Google Scholar] [CrossRef] [PubMed]
- Mayorga, M.; Romero-Salazar, L.; Rubi, J. Stochastic model for the dynamics of interacting Brownian particles. Phys. A 2002, 307, 297–314. [Google Scholar] [CrossRef]
- Savel’ev, S.; Marchesoni, F.; Taloni, A.; Nori, F. Diffusion of interacting Brownian particles: Jamming and anomalous diffusion. Phys. Rev. E 2006, 74, 021119. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.J.; Miller, J.F. Cartesian Genetic Programming: Why no bloat? In European Conference on Genetic Programming—EuroGP LNCS 8599; Nicolau, M., Krawiec, K., Heywood, M.I., Castelli, M., García-Sánchez, P., Merelo, J.J., Rivas Santos, V.M., Sim, K., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2014; pp. 222–233. [Google Scholar]
- Scheffer, M.; Carpenter, S.R.; Lenton, T.M.; Bascompte, J.; Brock, W.; Dakos, V.; van de Koppel, J.; van de Leemput, I.A.; Levin, S.A.; van Nes, E.H.; et al. Anticipating critical transitions. Science 2012, 338, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Clark, J. Functionality, Complexity, and Approaches to Assessment of Resilience under Constrained Energy and Information. Ph.D. Thesis, Air Force Institute of Technology, Wright-Patterson AFB, Greene, OH, USA, 2015. AFIT-ENV-DS-15-M-159. [Google Scholar]
- Lloyd, S.; Pagels, H. Complexity as thermodynamic depth. Ann. Phys. 1988, 188, 186–213. [Google Scholar] [CrossRef]
- Corning, P.A. Complexity is just a word! Technol. Forecast. Soc. Chang. 1998, 58, 1–4. [Google Scholar] [CrossRef]
- Crutchfield, J.P.; Shalizi, C.R. Thermodynamic depth of causal states: When paddling around in Occam’s pool shallowness is a virtue. Phys. Rev. E 1999, 59, 275–283. [Google Scholar] [CrossRef]
- Li, W. On the relationship between complexity and entropy for Markov chains and regular languages. Complexity 1991, 5, 381–399. [Google Scholar]
- Bar-Yam, Y. Multiscale Complexity/Entropy. Adv. Complex Syst. 2004, 7, 47–63. [Google Scholar] [CrossRef]
- Chaisson, E.J. Energy rate density as a complexity metric and evolutionary driver. Complexity 2011, 16, 27–40. [Google Scholar] [CrossRef]
- INCOSE. INCOSE Resilient Systems Working Group (RSWG) Charter. 2011. Available online: http://www.incose.org/docs/default-source/wgcharters/resilient-systems.pdf?sfvrsn=6 (accessed on 2 July 2017).
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seffens, W. Anomalous Diffusion within the Transcriptome as a Bio-Inspired Computing Framework for Resilience. Computation 2017, 5, 32. https://doi.org/10.3390/computation5030032
Seffens W. Anomalous Diffusion within the Transcriptome as a Bio-Inspired Computing Framework for Resilience. Computation. 2017; 5(3):32. https://doi.org/10.3390/computation5030032
Chicago/Turabian StyleSeffens, William. 2017. "Anomalous Diffusion within the Transcriptome as a Bio-Inspired Computing Framework for Resilience" Computation 5, no. 3: 32. https://doi.org/10.3390/computation5030032



