Cellular Uptake of Tile-Assembled DNA Nanotubes
Abstract
:1. Introduction
2. Results and Discussion
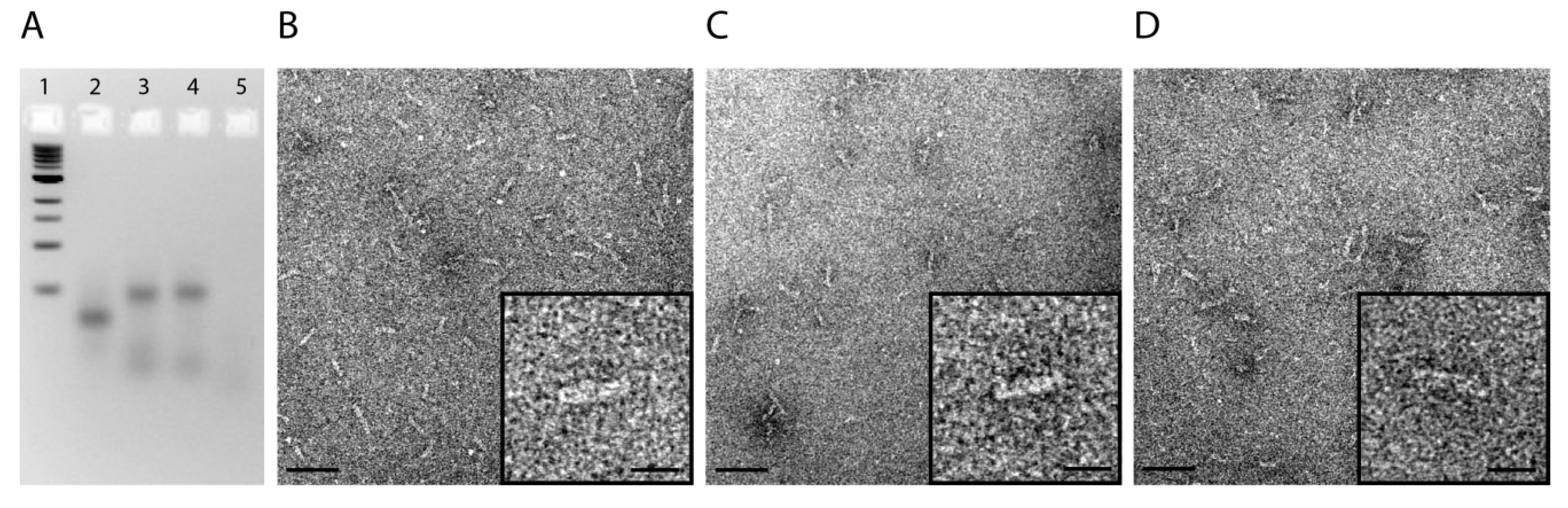
2.1. Design and Self-Assembly of Six-Helix DNA Nanotubes
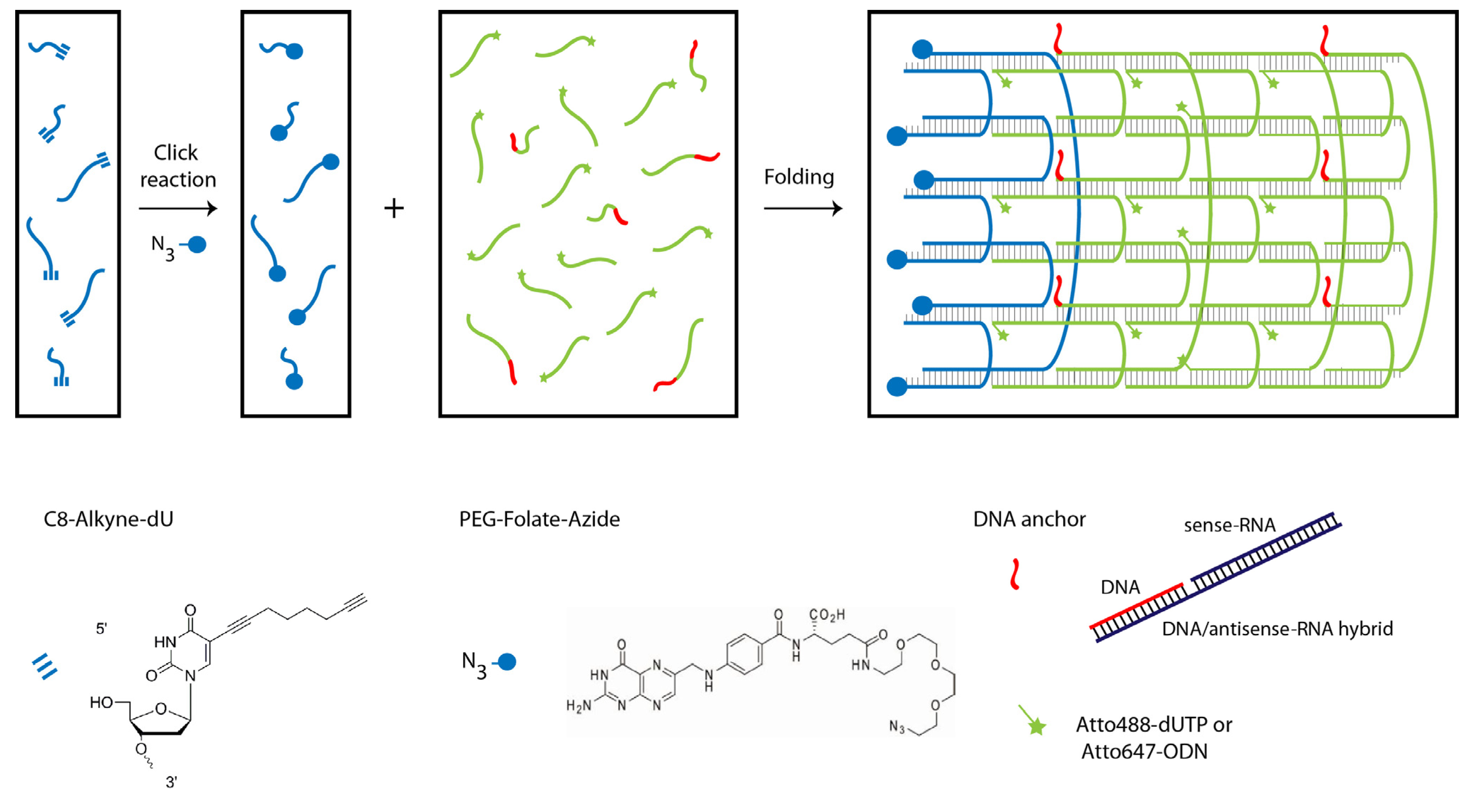
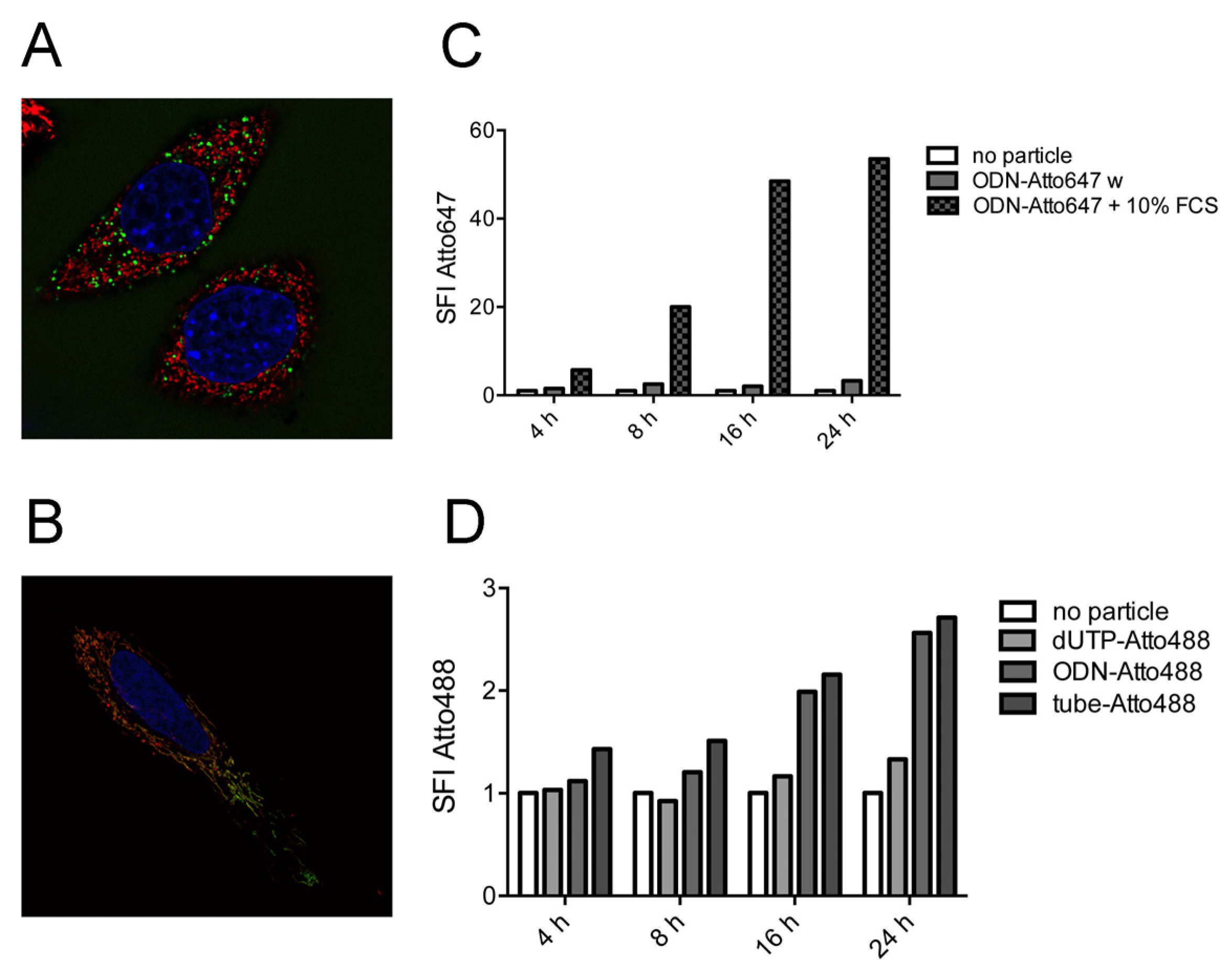
2.2. Tubule-Like Tile-Assembled DNA Nanostructures Are Delivered to the Endosome of HeLa Cells Independently of Folic Acid and Are Not Capable of Releasing siRNA into the Cytosol

2.3. Stability of DNA Nanotubes Differs in Various Conditions In Vitro


2.4. Strong Extra-Endosomal Uptake Can Be Feigned by Dye Cleavage
2.5. Single-Stranded DNA Molecules, But Not Deoxynucleotide Triphosphates, Are Internalized at Similar Levels as the Tile-Assembled Nanotube Structures
3. Experimental Section
3.1. DNA Nanotube Design
3.2. Folate Conjugation and Characterization of Oligonucleotides
3.3. Dye Labeling of DNA Nanotubes
3.4. DNA Nanotube Assembly and Purification
3.5. Gel Electrophoresis and Transmission Electron Microscopy
3.6. Stability of DNA Nanotubes
3.7. Cell Culture Experiments
3.8. Flow Cytometry and Confocal Fluorescence Microscopy
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bareford, L.M.; Swaan, P.W. Endocytic mechanisms for targeted drug delivery. Adv. Drug Deliv. Rev. 2007, 59, 748–758. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Schüller, V.; Engst, C.; Radler, J.; Liedl, T. Nucleic acid nanostructures for biomedical applications. Nanomedicine 2013, 8, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Harada, A.; Nagasaki, Y. Block copolymer micelles for drug delivery: Design, characterization and biological significance. Adv. Drug Deliv. Rev. 2001, 47, 113–131. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.D. Cationic liposomes for gene therapy. Angew. Chem. Int. Ed. 1998, 37, 1768–1785. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Therapeutic potential of aptamer-siRNA conjugates for treatment of HIV-1. BioDrugs 2012, 26, 393–400. [Google Scholar] [PubMed]
- Lee, H.; Lytton-Jean, A.K.; Chen, Y.; Love, K.T.; Park, A.I.; Karagiannis, E.D.; Sehgal, A.; Querbes, W.; Zurenko, C.S.; Jayaraman, M.; et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012, 7, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pei, H.; Zhu, B.; Liang, L.; Wei, M.; He, Y.; Chen, N.; Li, D.; Huang, Q.; Fan, C. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory cpg oligonucleotides. ACS Nano 2011, 5, 8783–8789. [Google Scholar] [CrossRef] [PubMed]
- Schüller, V.J.; Heidegger, S.; Sandholzer, N.; Nickels, P.C.; Suhartha, N.A.; Endres, S.; Bourquin, C.; Liedl, T. Cellular immunostimulation by cpg-sequence-coated DNA origami structures. ACS Nano 2011, 5, 9696–9702. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-X.; Shaw, A.; Zeng, X.; Benson, E.; Nyström, A.M.; Högberg, B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 2012, 6, 8684–8691. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Song, C.; Nangreave, J.; Liu, X.; Lin, L.; Qiu, D.; Wang, Z.-G.; Zou, G.; Liang, X.; Yan, H.; et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403. [Google Scholar] [CrossRef] [PubMed]
- Voigt, N.V.; Torring, T.; Rotaru, A.; Jacobsen, M.F.; Ravnsbaek, J.B.; Subramani, R.; Mamdouh, W.; Kjems, J.; Mokhir, A.; Besenbacher, F.; et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 2010, 5, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Hogberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Perrault, S.D.; Shih, W.M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 2014, 8, 5132–5140. [Google Scholar] [CrossRef] [PubMed]
- Mikkilä, J.; Eskelinen, A.-P.; Niemelä, E.H.; Linko, V.; Frilander, M.J.; Törmä, P.; Kostiainen, M.A. Virus-encapsulated DNA origami nanostructures for cellular delivery. Nano Lett. 2014, 14, 2196–2200. [Google Scholar] [CrossRef] [PubMed]
- Okholm, A.H.; Nielsen, J.S.; Vinther, M.; Sorensen, R.S.; Schaffert, D.; Kjems, J. Quantification of cellular uptake of DNA nanostructures by qPCR. Methods 2014, 67, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Liu, H.; Chen, Y.; Mao, C. DNA nanotubes as combinatorial vehicles for cellular delivery. Biomacromolecules 2008, 9, 3039–3043. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.A.; Jang, B.; Jeong, E.H.; Jeong, H.; Lee, H. Self-assembled DNA nanostructures prepared by rolling circle amplification for the delivery of sirna conjugates. Chem. Comm. 2014, 50, 13049–13051. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.; Wickham, S.F.; Shih, W.M.; Perrault, S.D. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Construction of three-dimensional stick figures from branched DNA. DNA Cell Biol. 1991, 10, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Erben, C.M.; Goodman, R.P.; Turberfield, A.J. Single-molecule protein encapsulation in a rigid DNA cage. Angew. Chem. Int. Ed. 2006, 45, 7414–7417. [Google Scholar] [CrossRef]
- Shih, W.M.; Quispe, J.D.; Joyce, G.F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 2004, 427, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Dai, M.; Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 2012, 485, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Ong, L.L.; Shih, W.M.; Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 2012, 338, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Hariadi, R.F.; Sahu, S.; Choi, H.M.; Park, S.H.; Labean, T.H.; Reif, J.H. Programming DNA tube circumferences. Science 2008, 321, 824–826. [Google Scholar] [CrossRef] [PubMed]
- Schiffels, D.; Liedl, T.; Fygenson, D.K. Nanoscale structure and microscale stiffness of DNA nanotubes. ACS Nano 2013, 7, 6700–6710. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.G.; Dietz, H. Magnesium-free self-assembly of multi-layer DNA objects. Nat. Comm. 2012, 3. [Google Scholar] [CrossRef]
- Di Michele, L.; Mognetti, B.M.; Yanagishima, T.; Varilly, P.; Ruff, Z.; Frenkel, D.; Eiser, E. Effect of inert tails on the thermodynamics of DNA hybridization. J. Am. Chem. Soc. 2014, 136, 6538–6541. [Google Scholar] [CrossRef] [PubMed]
- Kobold, S.; Steffen, J.; Chaloupka, M.; Grassmann, S.; Henkel, J.; Castoldi, R.; Zeng, Y.; Chmielewski, M.; Schmollinger, J.C.; Schnurr, M.; et al. Selective bispecific T cell recruiting antibody enhances anti-tumor activity of adoptive T cell transfer. J. Natl. Cancer Inst. 2014, 107. [Google Scholar] [CrossRef]
- Engels, B.; Cam, H.; Schuler, T.; Indraccolo, S.; Gladow, M.; Baum, C.; Blankenstein, T.; Uckert, W. Retroviral vectors for high-level transgene expression in T lymphocytes. Human Gene Ther. 2003, 14, 1155–1168. [Google Scholar] [CrossRef]
- Besch, R.; Poeck, H.; Hohenauer, T.; Senft, D.; Hacker, G.; Berking, C.; Hornung, V.; Endres, S.; Ruzicka, T.; Rothenfusser, S.; et al. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J. Clin. Invest. 2009, 119, 2399–2411. [Google Scholar] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kocabey, S.; Meinl, H.; MacPherson, I.S.; Cassinelli, V.; Manetto, A.; Rothenfusser, S.; Liedl, T.; Lichtenegger, F.S. Cellular Uptake of Tile-Assembled DNA Nanotubes. Nanomaterials 2015, 5, 47-60. https://doi.org/10.3390/nano5010047
Kocabey S, Meinl H, MacPherson IS, Cassinelli V, Manetto A, Rothenfusser S, Liedl T, Lichtenegger FS. Cellular Uptake of Tile-Assembled DNA Nanotubes. Nanomaterials. 2015; 5(1):47-60. https://doi.org/10.3390/nano5010047
Chicago/Turabian StyleKocabey, Samet, Hanna Meinl, Iain S. MacPherson, Valentina Cassinelli, Antonio Manetto, Simon Rothenfusser, Tim Liedl, and Felix S. Lichtenegger. 2015. "Cellular Uptake of Tile-Assembled DNA Nanotubes" Nanomaterials 5, no. 1: 47-60. https://doi.org/10.3390/nano5010047



