Characterization and Discrimination of Gram-Positive Bacteria Using Raman Spectroscopy with the Aid of Principal Component Analysis
Abstract
:1. Introduction
2. Results and Discussion
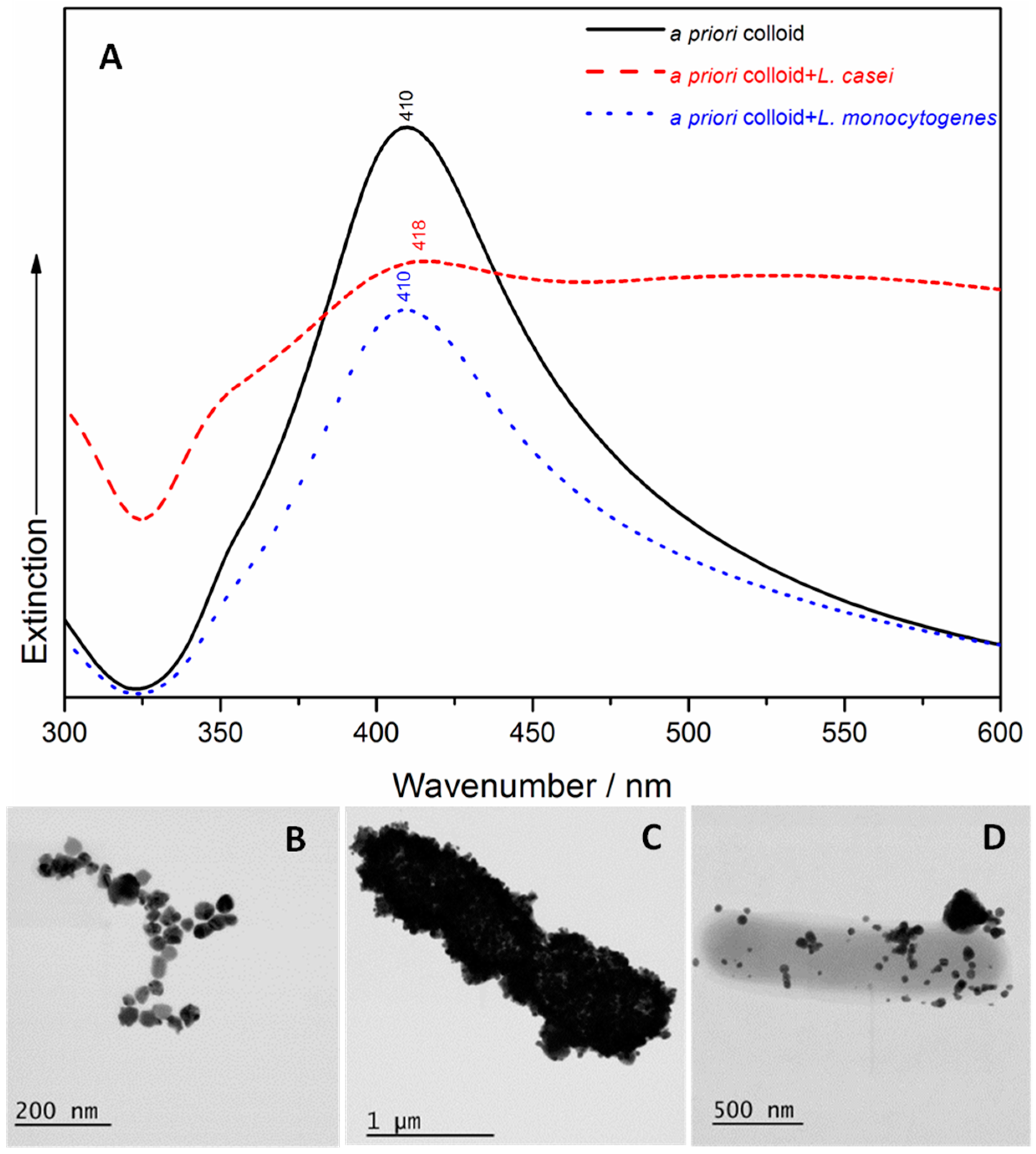
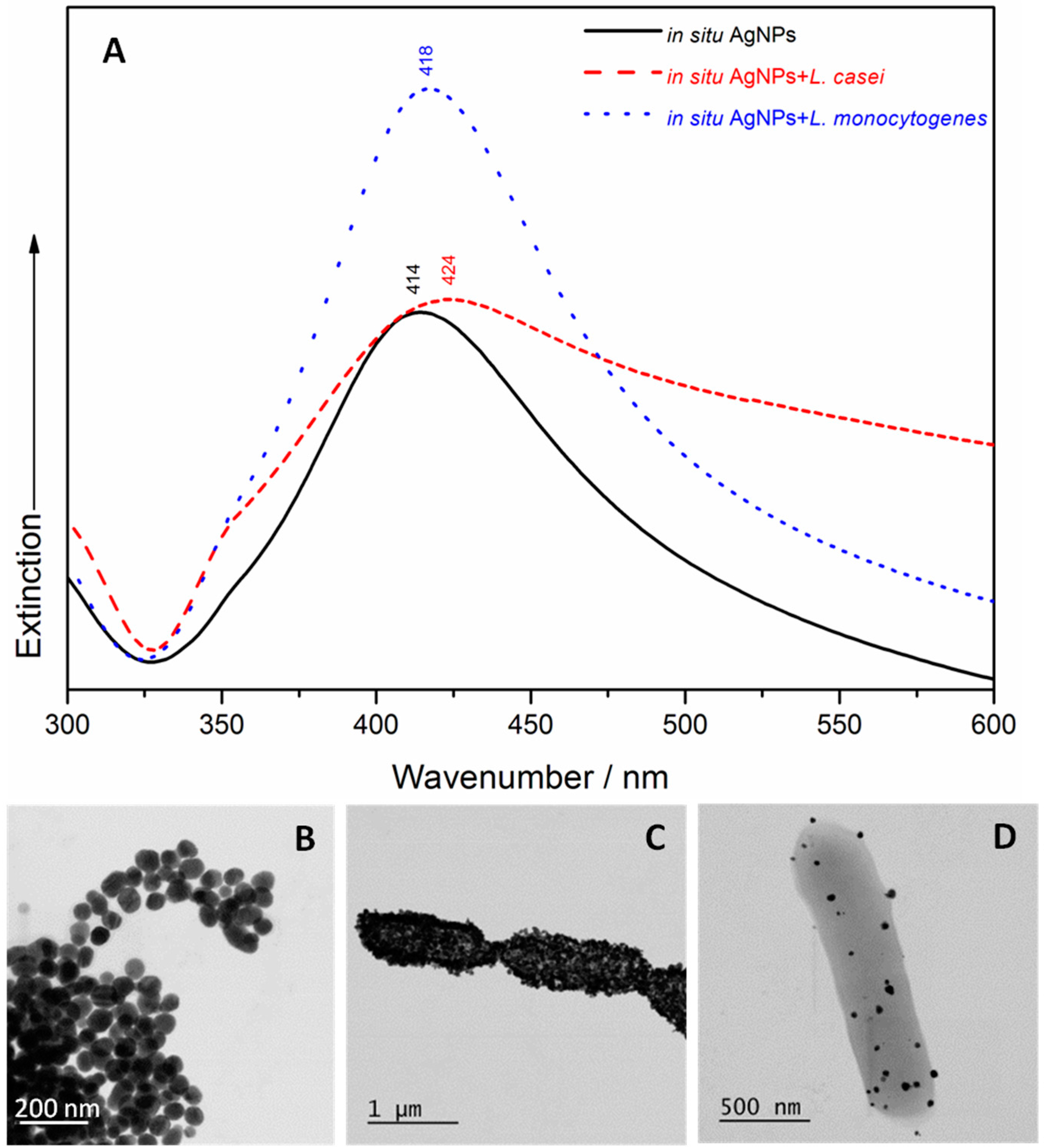
2.1. UV/Visible Spectroscopy and Scanning Transmission Electron Microscopy (STEM)
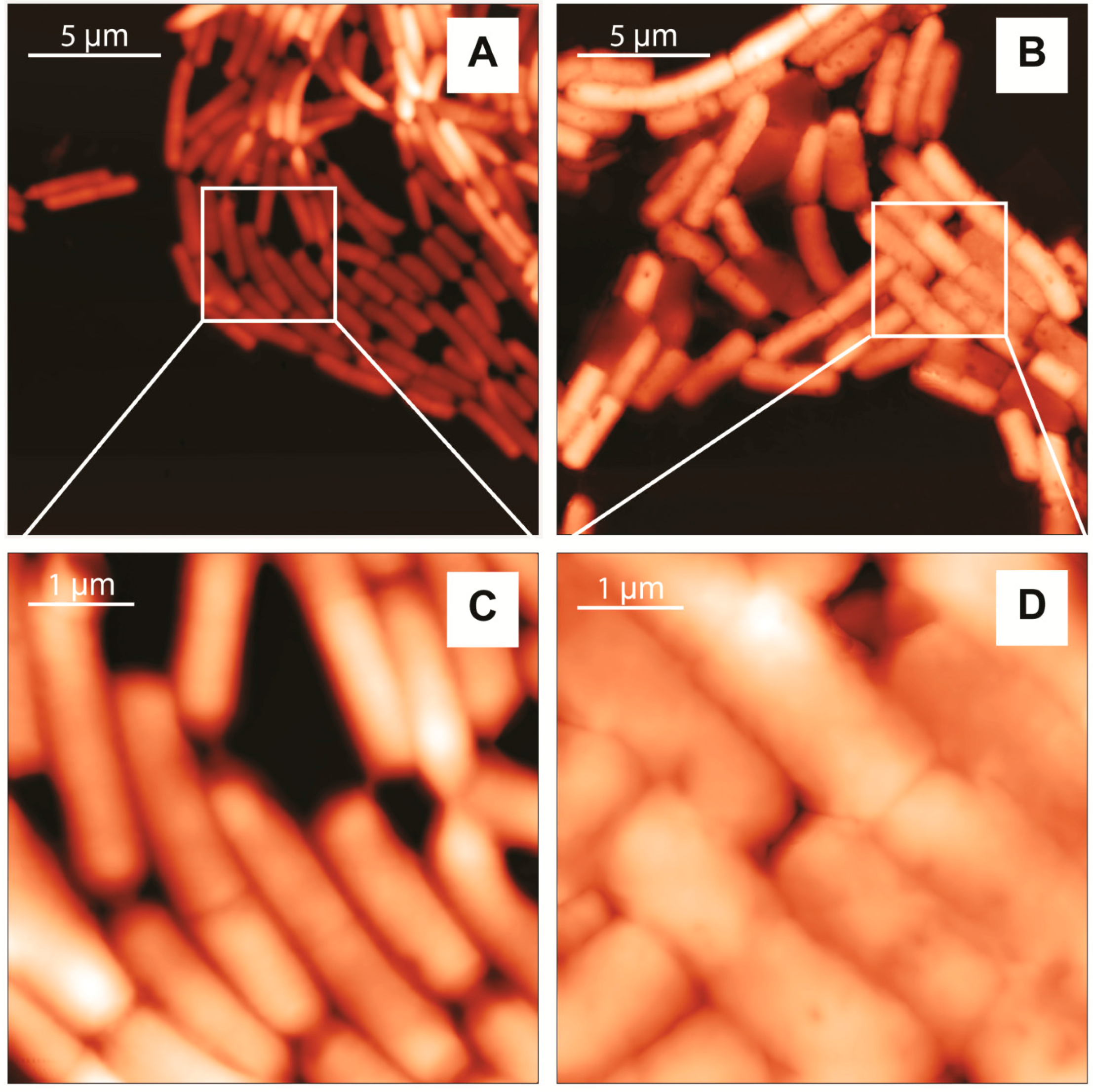
2.2. Atomic Force Microscopy (AFM) Characterization
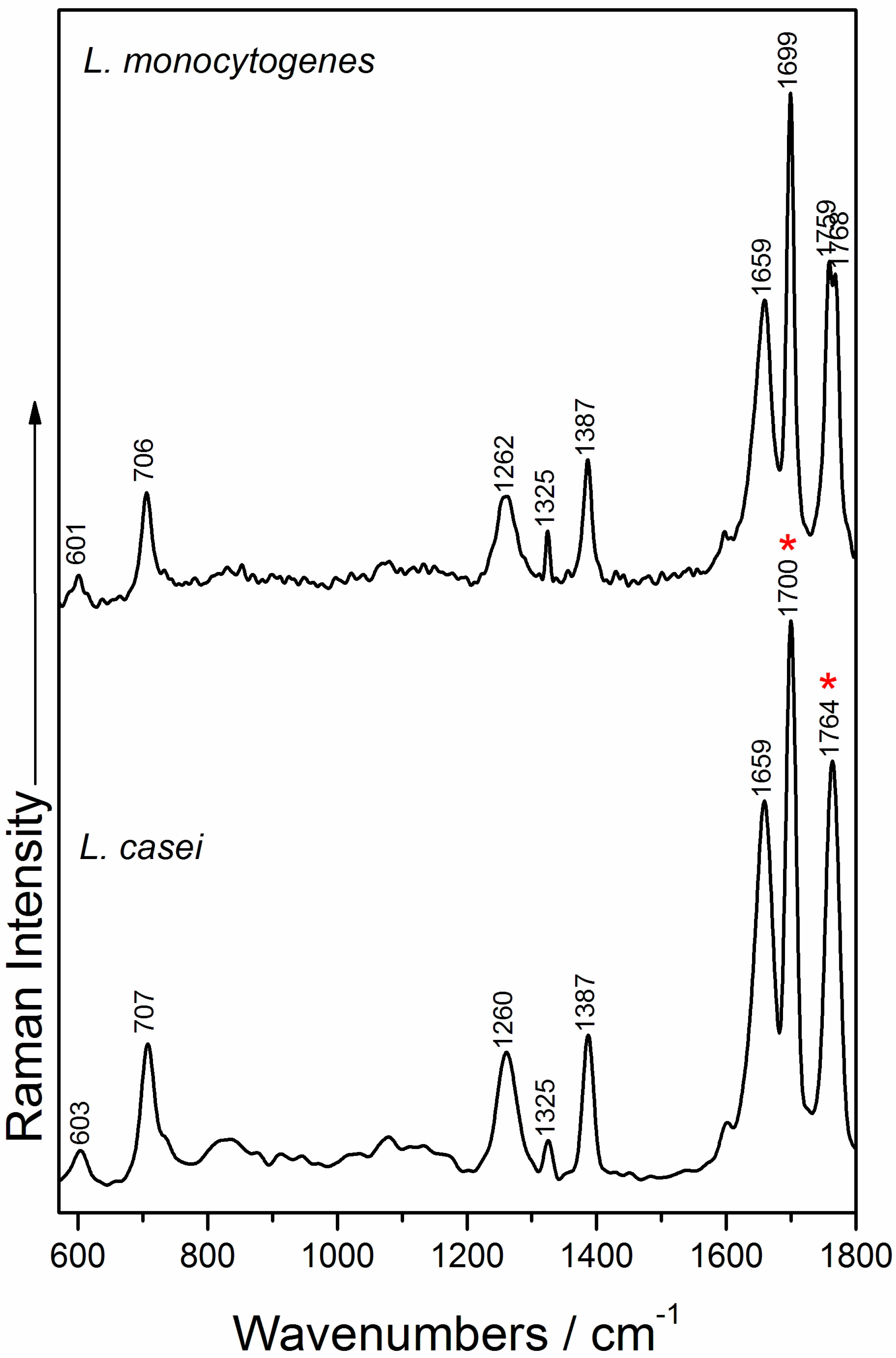
2.3. Raman Measurements and Analysis
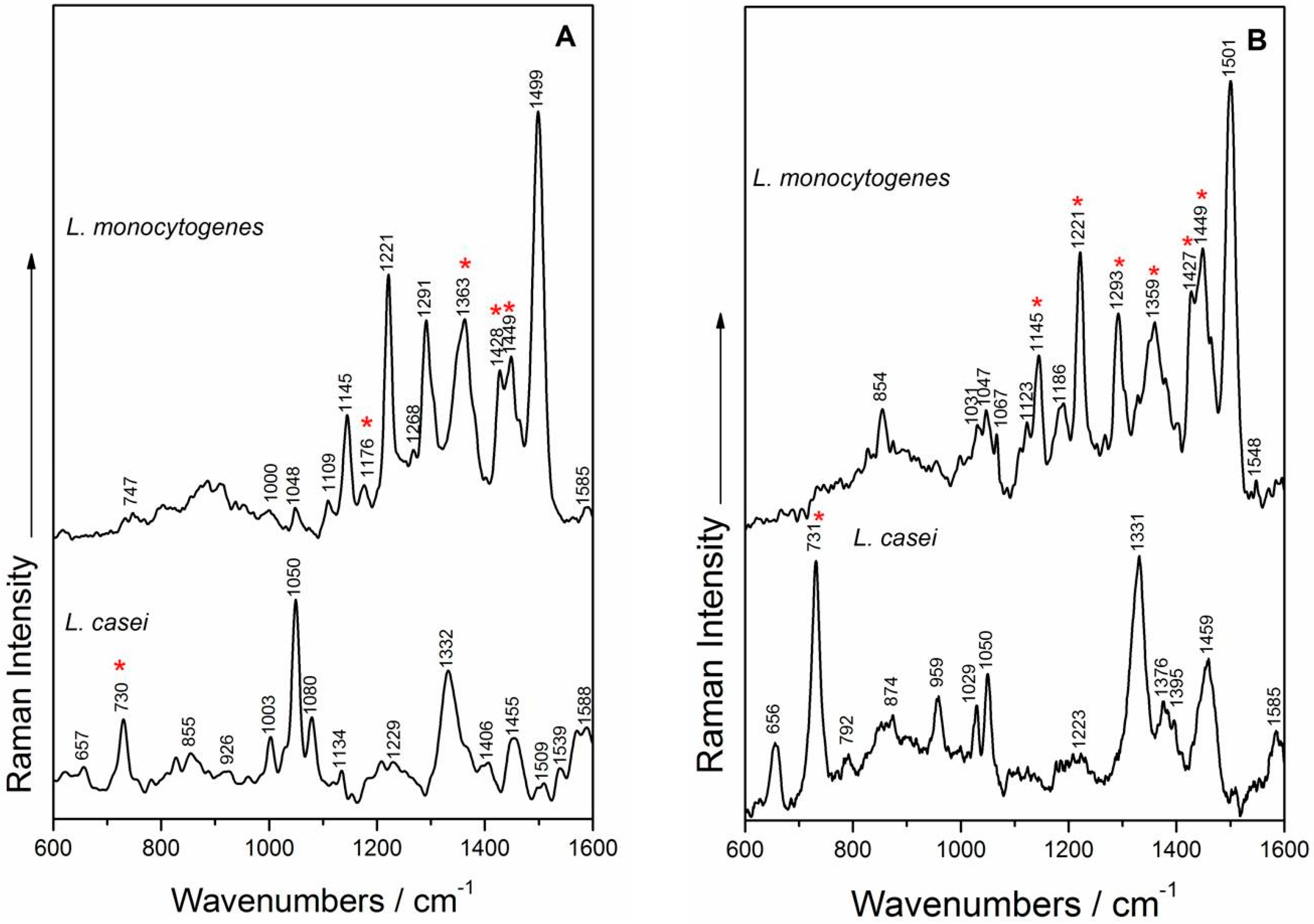
2.4. SERS Measurements and Analysis
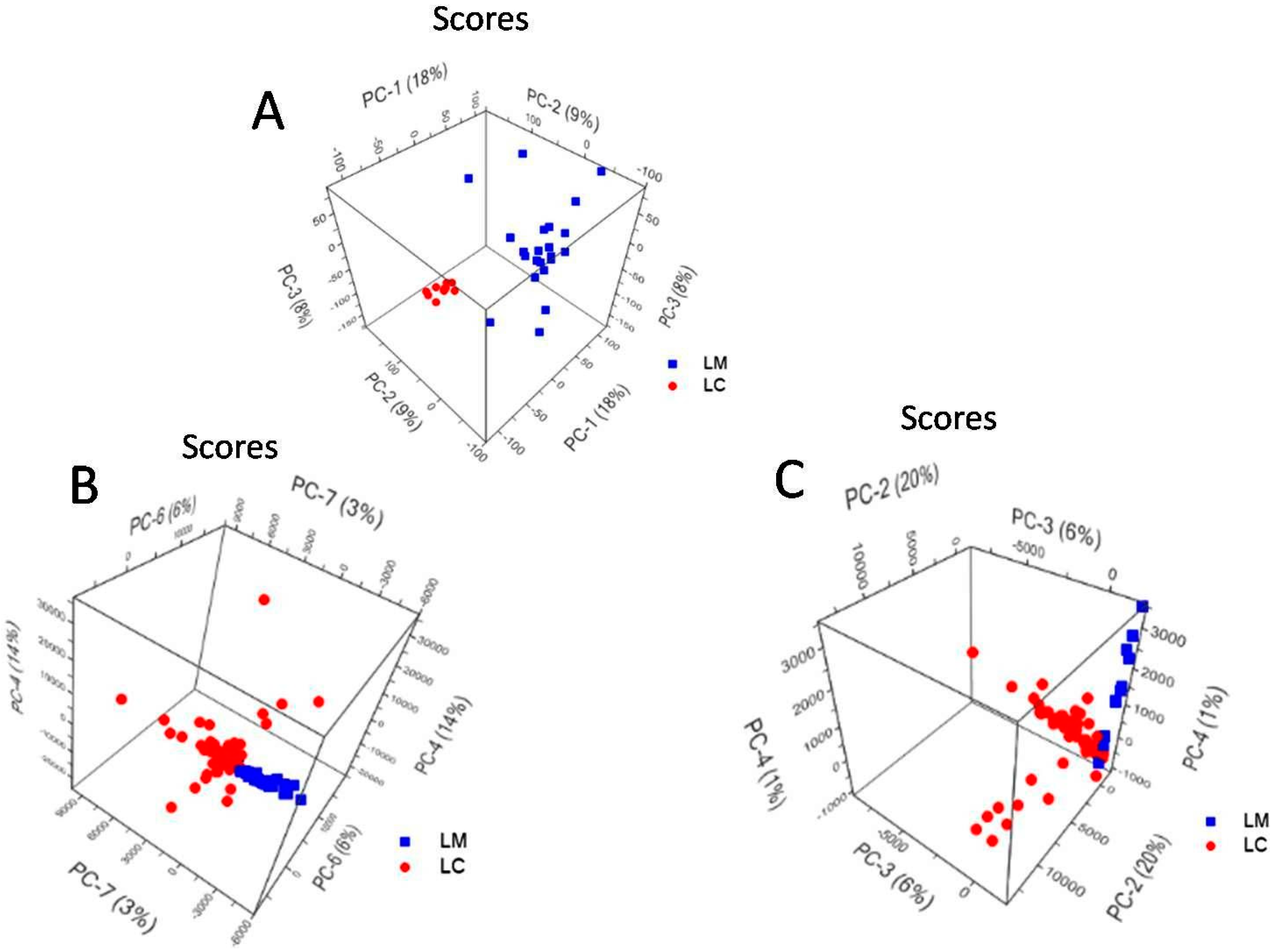
2.5. Principal Component Analysis of L. casei and L. monocytogenes Spectra
3. Materials and Methods
3.1. Bacteria Cultivation
3.2. A Priori Colloid Synthesis and In Situ NPs Synthesis
3.3. Raman and SERS Measurement Details
3.3.1. Instrumentation
3.3.2. Sample Preparation
3.3.3. Spectra Acquisition Details
3.4. UV Characterization
3.5. STEM Measurements
3.6. Computational Details
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cruz, P.; Mehretu, A.M.; Buttner, M.P.; Trice, T.; Howard, K.M. Development of a polymerase chain reaction assay for the rapid detection of the oral pathogenic bacterium, Selenomonas noxia. BMC Oral Health 2015, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Cho, J.-C. Simple and rapid detection of Listeria monocytogenes in fruit juice by real-time PCR without enrichment culture. Food Control 2010, 21, 1419–1423. [Google Scholar] [CrossRef]
- O’Grady, J.; Ruttledge, M.; Sedano-Balbas, S.; Smith, T.J.; Barry, T.; Maher, M. Rapid detection of Listeria monocytogenes in food using culture enrichment combined with real-time PCR. Food Microbiol. 2009, 26, 4–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Lu, L.; Pan, Y.; Sun, X.; Hwang, C.-A.; Zhao, Y.; Wu, V.C.H. Rapid detection and differentiation of Listeria monocytogenes and Listeria species in deli meats by a new multiplex PCR method. Food Control 2015, 52, 78–84. [Google Scholar] [CrossRef]
- Mendonca, M.; Conrad, N.L.; Conceicao, F.R.; Moreira, A.N.; da Silva, W.P.; Aleixo, J.A.; Bhunia, A.K. Highly specific fiber optic immunosensor coupled with immunomagnetic separation for detection of low levels of Listeria monocytogenes and L. ivanovii. BMC Microbiol. 2012, 12, 275. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Salazar, J.K. Culture-Independent Rapid Detection Methods for Bacterial Pathogens and Toxins in Food Matrices. Compr. Rev. Food Sci. Food. Saf. 2016, 15, 183–205. [Google Scholar] [CrossRef]
- Davis, D.; Guo, X.; Musavi, L.; Lin, C.-S.; Chen, S.-H.; Wu, V.C.H. Gold Nanoparticle-Modified Carbon Electrode Biosensor for the Detection of Listeria monocytogenes. Ind. Biotechnol. 2013, 9, 31–36. [Google Scholar] [CrossRef]
- Cho, I.H.; Irudayaraj, J. Lateral-flow enzyme immunoconcentration for rapid detection of Listeria monocytogenes. Anal. Bioanal. Chem. 2013, 405, 3313–3319. [Google Scholar] [CrossRef] [PubMed]
- Välimaa, A.-L.; Tilsala-Timisjärvi, A.; Virtanen, E. Rapid detection and identification methods for Listeria monocytogenes in the food chain—A review. Food Control 2015, 55, 103–114. [Google Scholar] [CrossRef]
- Troy, D.J.; Ojha, K.S.; Kerry, J.P.; Tiwari, B.K. Sustainable and consumer-friendly emerging technologies for application within the meat industry: An overview. Meat Sci. 2016, 120, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Mobili, P.; Londero, A.; De Antoni, G.; Gomez-Zavaglia, A. Multivariate analysis of Raman spectra applied to microbiology: Discrimination of microorganisms at the species level. Rev. Mex. Fis. 2010, 56, 378–385. [Google Scholar]
- Mircescu, N.E.; Zhou, H.; Leopold, N.; Chis, V.; Ivleva, N.P.; Niessner, R.; Wieser, A.; Haisch, C. Towards a receptor-free immobilization and SERS detection of urinary tract infections causative pathogens. Anal. Bioanal. Chem. 2014, 406, 3051–3058. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Hu, Z.; Mustapha, A.; Lin, M. Rapid detection of food- and waterborne bacteria using surface-enhanced Raman spectroscopy coupled with silver nanosubstrates. Appl. Microbiol. Biotechnol. 2011, 92, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Oust, A.; Moretro, T.; Naterstad, K.; Sockalingum, G.D.; Adt, I.; Manfait, M.; Kohler, A. Fourier transform Infrared and Raman spectroscopy for characterization of Listeria monocytogenes strains. Appl. Environ. Microbiol. 2006, 72, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Beuchat, L.R.; Doyle, M.P.; Montville, T.J. Food Microbiology: Fundamentals and Frontiers, 2nd ed.; ASM Press: Washington, DC, USA, 2001. [Google Scholar]
- Mungroo, N.A.; Oliveira, G.; Neethirajan, S. SERS based point-of-care detection of food-borne pathogens. Microchim. Acta 2016, 183, 697–707. [Google Scholar] [CrossRef]
- Craig, A.P.; Franca, A.S.; Irudayaraj, J. Surface-enhanced Raman spectroscopy applied to food safety. Ann. Rev. Food Sci. 2013, 4, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, L.; Jurasekova, Z.; Domingo, C.; Pérez-Méndez, M.; Leyton, P.; Campos-Vallette, M.; Garcia-Ramos, J.V.; Sanchez-Cortes, S. Importance of Metal-Adsorbate Interactions for the Surface-enhanced Raman Scattering of Molecules Adsorbed on Plasmonic Nanoparticles. Plasmonics 2007, 2, 147–156. [Google Scholar] [CrossRef]
- Leopold, N.; Lendl, B. A new method for fast preparation of highly surface-enhanced Raman scattering (SERS) active silver colloids at room temperature by reduction of silver nitrate with hydroxylamine hydrochloride. J. Phys. Chem. B 2003, 107, 5723–5727. [Google Scholar] [CrossRef]
- Tódor, I.S.; Szabó, L.; Marişca, O.T.; Chiş, V.; Leopold, N. Gold nanoparticle assemblies of controllable size obtained by hydroxylamine reduction at room temperature. J. Nanopart. Res. 2014, 16, 2740. [Google Scholar] [CrossRef]
- Leopold, N.; Chiş, V.; Mircescu, N.E.; Marişca, O.T.; Buja, O.M.; Leopold, L.F.; Socaciu, C.; Braicu, C.; Irimie, A.; Berindan-Neagoe, I. One step synthesis of SERS active colloidal gold nanoparticles by reduction with polyethylene glycol. Colloids Surf. A Physicochem. Eng. Asp. 2013, 436, 133–138. [Google Scholar] [CrossRef]
- Buja, O.M.; Mircescu, N.E.; Leopold, N. Raman Scattering Enhancement of Peg Coated Gold Nanoparticles of Defined Size. J. Appl. Spectrosc. 2014, 81, 411–415. [Google Scholar] [CrossRef]
- Marisca, O.; Kantner, K.; Pfeiffer, C.; Zhang, Q.; Pelaz, B.; Leopold, N.; Parak, W.; Rejman, J. Comparison of the in Vitro Uptake and Toxicity of Collagen- and Synthetic Polymer-Coated Gold Nanoparticles. Nanomaterials 2015, 5, 1418–1430. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, M.; Wachsmann-Hogiu, S. Plasmonic Nanostructures for Bioanalytical Applications of SERS. In Plasmonics in Biology and Medicine Xiii; VoDinh, T., Lakowicz, J.R., Ho, H.P.A., Ray, K., Eds.; SPIE: Bellingham, WA, USA, 2016; Volume 9724. [Google Scholar]
- Vishnupriya, S.; Chaudhari, K.; Jagannathan, R.; Pradeep, T. Single-Cell Investigations of Silver Nanoparticle–Bacteria Interactions. Part. Part. Syst. Charact. 2013, 30, 1056–1062. [Google Scholar] [CrossRef]
- Efrima, S.; Bronk, B.V. Silver Colloids Impregnating or Coating Bacteria. J. Phys. Chem. B 1998, 102, 5947–5950. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, D.; Ivleva, N.P.; Mircescu, N.E.; Niessner, R.; Haisch, C. SERS detection of bacteria in water by in situ coating with Ag nanoparticles. Anal. Chem. 2014, 86, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.B.; Yang, D.T.; Mircescu, N.; Ivleva, N.P.; Schwarzmeier, K.; Wieser, A.; Schubert, S.; Niessner, R.; Haisch, C. Surface-enhanced Raman scattering detection of bacteria on microarrays at single cell levels using silver nanoparticles. Microchim. Acta 2015, 182, 2259–2266. [Google Scholar] [CrossRef]
- Dina, N.E.; Zhou, H.; Colnita, A.; Leopold, N.; Szoke-Nagy, T.; Coman, C.; Haisch, C. Rapid single-cell detection and identification of pathogens by using surface-enhanced Raman spectroscopy. Analyst 2017, 142, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, D.; Ivleva, N.P.; Mircescu, N.E.; Schubert, S.; Niessner, R.; Wieser, A.; Haisch, C. Label-Free in Situ Discrimination of Live and Dead Bacteria by Surface-Enhanced Raman Scattering. Anal. Chem. 2015, 87, 6553–6561. [Google Scholar] [CrossRef] [PubMed]
- Dina, N.E.; Colnita, A.; Szoke-Nagy, T.; Porav, A.S. A Critical Review on Ultrasensitive, Spectroscopic-based Methods for High-throughput Monitoring of Bacteria during Infection Treatment. Crit. Rev. Anal. Chem. 2017, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, H.; Hu, Z.; Yu, G.; Yang, D.; Zhao, J. Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosens. Bioelectron. 2017, 94, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, M.; Snell, E.E. Cell Wall Composition of Lactic Acid Bacteria. J. Biol. Chem. 1960, 235, 1376–1382. [Google Scholar] [PubMed]
- Delcour, J.; Ferain, T.; Deghorain, M.; Palumbo, E.; Hols, P. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie van Leeuwenhoek J. Microb. 1999, 76, 159–184. [Google Scholar] [CrossRef]
- Vadillo-Rodríguez, V.; Busscher, H.J.; Norde, W.; de Vries, J.; van der Mei, H.C. Dynamic Cell Surface Hydrophobicity of Lactobacillus Strains with and without Surface Layer Proteins. J. Bacteriol. 2004, 186, 6647–6650. [Google Scholar] [CrossRef] [PubMed]
- Van der Mei, H.C.; de Vries, J.; Busscher, H.J. X-ray photoelectron spectroscopy for the study of microbial cell surfaces. Surf. Sci. Rep. 2000, 39, 1–24. [Google Scholar] [CrossRef]
- Goldfine, H.; Shen, H. Listeria Monocytogenes: Pathogenesis and Host Response; Springer Science: New York, NY, USA, 2007. [Google Scholar]
- Schär-Zammaretti, P.; Ubbink, J. The Cell Wall of Lactic Acid Bacteria: Surface Constituents and Macromolecular Conformations. Biophys. J. 2003, 85, 4076–4092. [Google Scholar] [CrossRef]
- Mobili, P.; Araujo-Andrade, C.; Londero, A.; Frausto-Reyes, C.; Tzonchev, R.I.; De Antoni, G.L.; Gomez-Zavaglia, A. Development of a method based on chemometric analysis of Raman spectra for the discrimination of heterofermentative lactobacilli. J. Dairy Res. 2011, 78, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Guicheteau, J.; Christesen, S.; Emge, D.; Tripathi, A. Bacterial mixture identification using Raman and surface-enhanced Raman chemical imaging. J. Raman Spectrosc. 2010, 41, 1632–1637. [Google Scholar] [CrossRef]
- Gaus, K.; Rosch, P.; Petry, R.; Peschke, K.D.; Ronneberger, O.; Burkhardt, H.; Baumann, K.; Popp, J. Classification of lactic acid bacteria with UV-resonance Raman spectroscopy. Biopolymers 2006, 82, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Gerbino, E.; Tymczyszyn, E.; Gomez-Zavaglia, A. Applications of Infrared and Raman Spectroscopies to Probiotic Investigation. Foods 2015, 4, 283–305. [Google Scholar] [CrossRef] [PubMed]
- Uusitalo, S.; Kogler, M.; Valimaa, A.L.; Popov, A.; Ryabchikov, Y.; Kontturi, V.; Siitonen, S.; Petaja, J.; Virtanen, T.; Laitinen, R.; et al. Detection of Listeria innocua on roll-to-roll produced SERS substrates with gold nanoparticles. RSC Adv. 2016, 6, 62981–62989. [Google Scholar] [CrossRef]
- Stambach, N.R.; Carr, S.A.; Cox, C.R.; Voorhees, K.J. Rapid Detection of Listeria by Bacteriophage Amplification and SERS-Lateral Flow Immunochromatography. Viruses 2015, 7, 6631–6641. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Morgan, M.T.; Bhunia, A.K. Detection of low levels of Listeria monocytogenes cells by using a fiber-optic immunosensor. Appl. Environ. Microbiol. 2004, 70, 6138–6146. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.M.; Bhunia, A.K. Specific detection of cytopathogenic Listeria monocytogenes using a two-step method of immunoseparation and cytotoxicity analysis. J. Microbiol. Methods 2005, 60, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Ohk, S.H.; Koo, O.K.; Sen, T.; Yamamoto, C.M.; Bhunia, A.K. Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J. Appl. Microbiol. 2010, 109, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.K.; Heighton, L.; Xu, Y.; Nou, X.; Schmidt, W.F. Raman mapping of intact biofilms on stainless steel surfaces. Proc. SPIE 2016, 9864, 986415–986417. [Google Scholar]
- Jo, Y.; Jung, J.; Kim, M.H.; Park, H.; Kang, S.J.; Park, Y. Label-free identification of individual bacteria using Fourier transform light scattering. Opt. Express 2015, 23, 15792–15805. [Google Scholar] [CrossRef] [PubMed]
- Grow, A.E.; Wood, L.L.; Claycomb, J.L.; Thompson, P.A. New biochip technology for label-free detection of pathogens and their toxins. J. Microbiol. Methods 2003, 53, 221–233. [Google Scholar] [CrossRef]
- Hernandez-Mendoza, A.; Guzman-De-Peña, D.; González-Córdova, A.; Vallejo-Córdoba, B.; Garcia, H. In vivo assessment of the potential protective effect of Lactobacillus casei Shirota against aflatoxin B1. Dairy Sci. Technol. 2010, 90, 729–740. [Google Scholar] [CrossRef]
- Nicholas, R.; Dunton, P.; Tatham, A.; Fielding, L. The effect of ozone and open air factor on surface-attached and biofilm environmental Listeria monocytogenes. J. Appl. Microbiol. 2013, 115, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Mercanoglu, B.; Aykut Aytac, S.; Ali Ergun, M.; Tan, E. Isolation of Listeria monocytogenes by Immunomagnetic Separation and Atomic Force Microscopy. J. Microbiol. (Seoul) 2003, 41, 144–147. [Google Scholar]
- JGI Genome Portal. Available online: http://genome.jgi-psf.org/lacca/lacca.home.html (accessed on 30 June 2016).
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.P.; van den Braak, N.; Endtz, H.P.; Naumann, D.; Puppels, G.J. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 2002, 51, 255–271. [Google Scholar] [CrossRef]
- Lu, X.; Al-Qadiri, H.; Lin, M.; Rasco, B. Application of Mid-infrared and Raman Spectroscopy to the Study of Bacteria. Food Bioprocess. Technol. 2011, 4, 919–935. [Google Scholar] [CrossRef]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Vancanneyt, M.; De Vos, P.; Moens, L. Methods for extracting biochemical information from bacterial Raman spectra: Focus on a group of structurally similar biomolecules-fatty acids. Anal. Chim. Acta 2007, 603, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Saxena, A.; Song, B.; Ward, B.B.; Beveridge, T.J.; Myneni, S.C.B. Elucidation of Functional Groups on Gram-Positive and Gram-Negative Bacterial Surfaces Using Infrared Spectroscopy. Langmuir 2004, 20, 11433–11442. [Google Scholar] [CrossRef] [PubMed]
- Childs, W.C.; Taron, D.J.; Neuhaus, F.C. Biosynthesis of D-alanyl-lipoteichoic acid by Lactobacillus casei: Interchain transacylation of D-alanyl ester residues. J. Bacteriol. 1985, 162, 1191–1195. [Google Scholar] [PubMed]
- Kahraman, M.; Zamaleeva, A.I.; Fakhrullin, R.F.; Culha, M. Layer-by-layer coating of bacteria with noble metal nanoparticles for surface-enhanced Raman scattering. Anal. Bioanal. Chem. 2009, 395, 2559–2567. [Google Scholar] [CrossRef] [PubMed]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Walter, A.; Marz, A.; Schumacher, W.; Rosch, P.; Popp, J. Towards a fast, high specific and reliable discrimination of bacteria on strain level by means of SERS in a microfluidic device. Lab Chip 2011, 11, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, M.; Keseroglu, K.; Culha, M. On sample preparation for surface-enhanced Raman scattering (SERS) of bacteria and the source of spectral features of the spectra. Appl. Spectrosc. 2011, 65, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Ivleva, N.P.; Wagner, M.; Szkola, A.; Horn, H.; Niessner, R.; Haisch, C. Label-freein situ SERS imaging of biofilms. J. Phys. Chem. B 2010, 114, 10184–10194. [Google Scholar] [CrossRef] [PubMed]
- Zeiri, L.; Bronk, B.V.; Shabtai, Y.; Eichler, J.; Efrima, S. Surface-enhanced Raman spectroscopy as a tool for probing specific biochemical components in bacteria. Appl. Spectrosc. 2004, 58, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Premasiri, W.R.; Lee, J.C.; Sauer-Budge, A.; Théberge, R.; Costello, C.E.; Ziegler, L.D. The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS. Anal. Bioanal. Chem. 2016, 408, 4631–4647. [Google Scholar] [CrossRef] [PubMed]
- Ashton, L.; Lau, K.; Winder, C.L.; Goodacre, R. Raman spectroscopy: Lighting up the future of microbial identification. Future Microbiol. 2011, 6, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Prucek, R.; Ranc, V.; Kvitek, L.; Panacek, A.; Zboril, R.; Kolar, M. Reproducible discrimination between Gram-positive and Gram-negative bacteria using surface enhanced Raman spectroscopy with infrared excitation. Analyst 2012, 137, 2866–2870. [Google Scholar] [CrossRef] [PubMed]
- Vohnik, S.; Hanson, C.; Tuma, R.; Fuchs, J.A.; Woodward, C.; Thomas, G.J., Jr. Conformation, stability, and active-site cysteine titrations of Escherichia coli D26A thioredoxin probed by Raman spectroscopy. Protein Sci. 1998, 7, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Sivakesava, S.; Irudayaraj, J.; Ali, D. Simultaneous determination of multiple components in lactic acid fermentation using FT-MIR, NIR, and FT-Raman spectroscopic techniques. Process Biochem. 2001, 37, 371–378. [Google Scholar] [CrossRef]
- Wang, J.; Xie, X.; Feng, J.; Chen, J.C.; Du, X.J.; Luo, J.; Lu, X.; Wang, S. Rapid detection of Listeria monocytogenes in milk using confocal micro-Raman spectroscopy and chemometric analysis. Int. J. Food Microbiol. 2015, 204, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Vodnar, D.; Socaciu, C. Green tea increases the survival yield of Bifidobacteria in simulated gastrointestinal environment and during refrigerated conditions. Chem. Cent. J. 2012, 6, 61. [Google Scholar] [CrossRef] [PubMed]
| Bacteria Species | L. casei | L. monocytogenes |
|---|---|---|
| Dimension (µm) | ||
| Length | 2.01 ± 0.42 | 2.14 ± 0.33 |
| Width | 0.72 ± 0.11 | 1.12 ± 0.07 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colniță, A.; Dina, N.E.; Leopold, N.; Vodnar, D.C.; Bogdan, D.; Porav, S.A.; David, L. Characterization and Discrimination of Gram-Positive Bacteria Using Raman Spectroscopy with the Aid of Principal Component Analysis. Nanomaterials 2017, 7, 248. https://doi.org/10.3390/nano7090248
Colniță A, Dina NE, Leopold N, Vodnar DC, Bogdan D, Porav SA, David L. Characterization and Discrimination of Gram-Positive Bacteria Using Raman Spectroscopy with the Aid of Principal Component Analysis. Nanomaterials. 2017; 7(9):248. https://doi.org/10.3390/nano7090248
Chicago/Turabian StyleColniță, Alia, Nicoleta Elena Dina, Nicolae Leopold, Dan Cristian Vodnar, Diana Bogdan, Sebastian Alin Porav, and Leontin David. 2017. "Characterization and Discrimination of Gram-Positive Bacteria Using Raman Spectroscopy with the Aid of Principal Component Analysis" Nanomaterials 7, no. 9: 248. https://doi.org/10.3390/nano7090248