Radial Flow Assay Using Gold Nanoparticles and Rolling Circle Amplification to Detect Mercuric Ions
Abstract
:1. Introduction
2. Results and Discussion
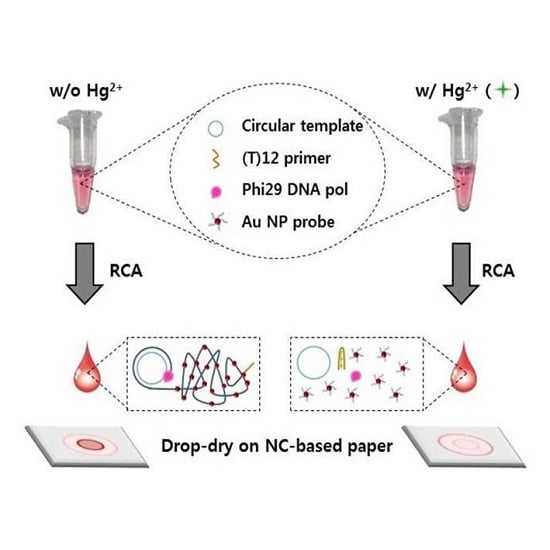
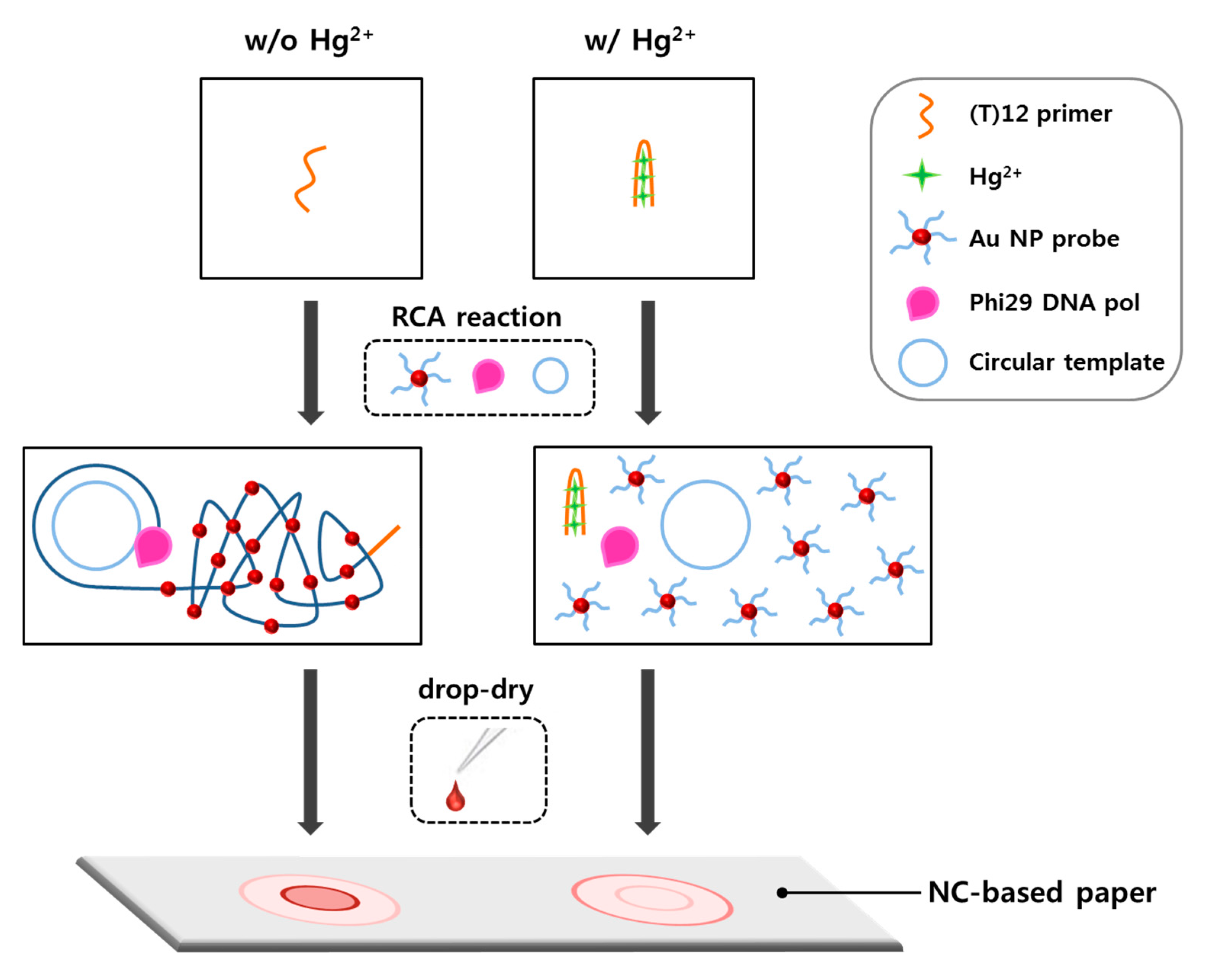
2.1. Design of Radial Flow Assay for Hg2+ Detection
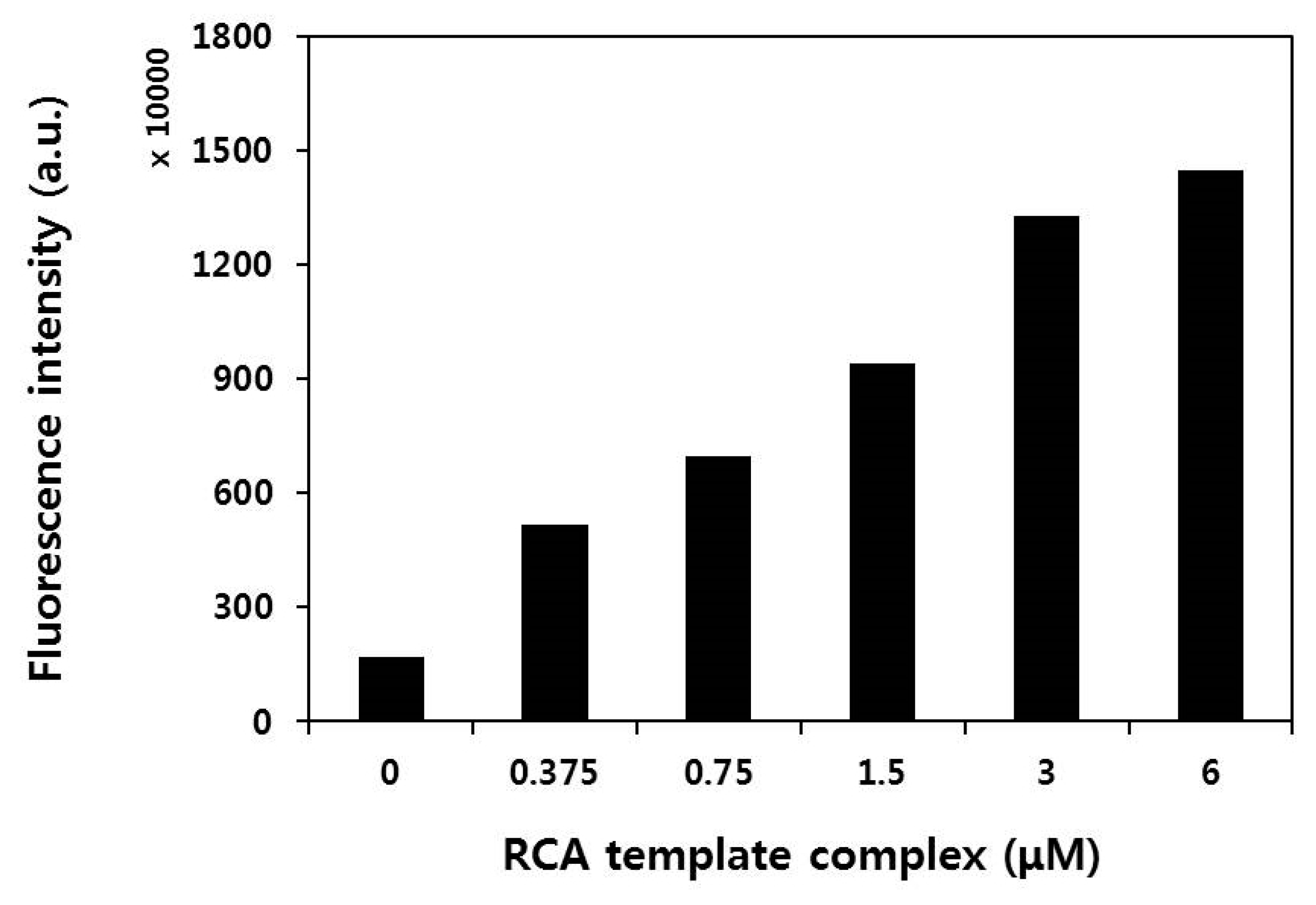
2.2. RCA Reaction Test Using Circular Template–(T)12 Primer Complexes
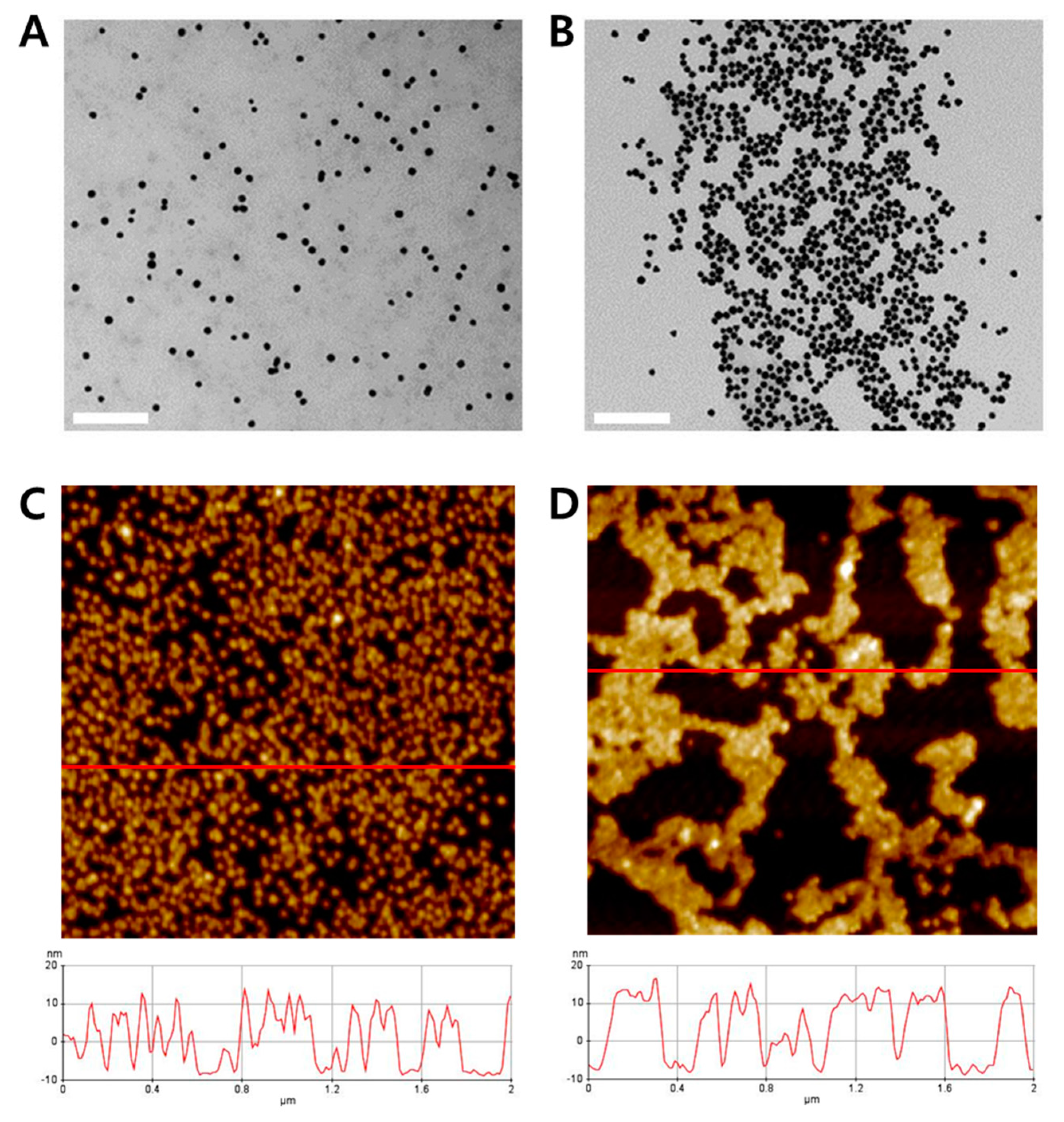
2.3. Formation of DNA Coil-AuNP Complexes by RCA Reaction
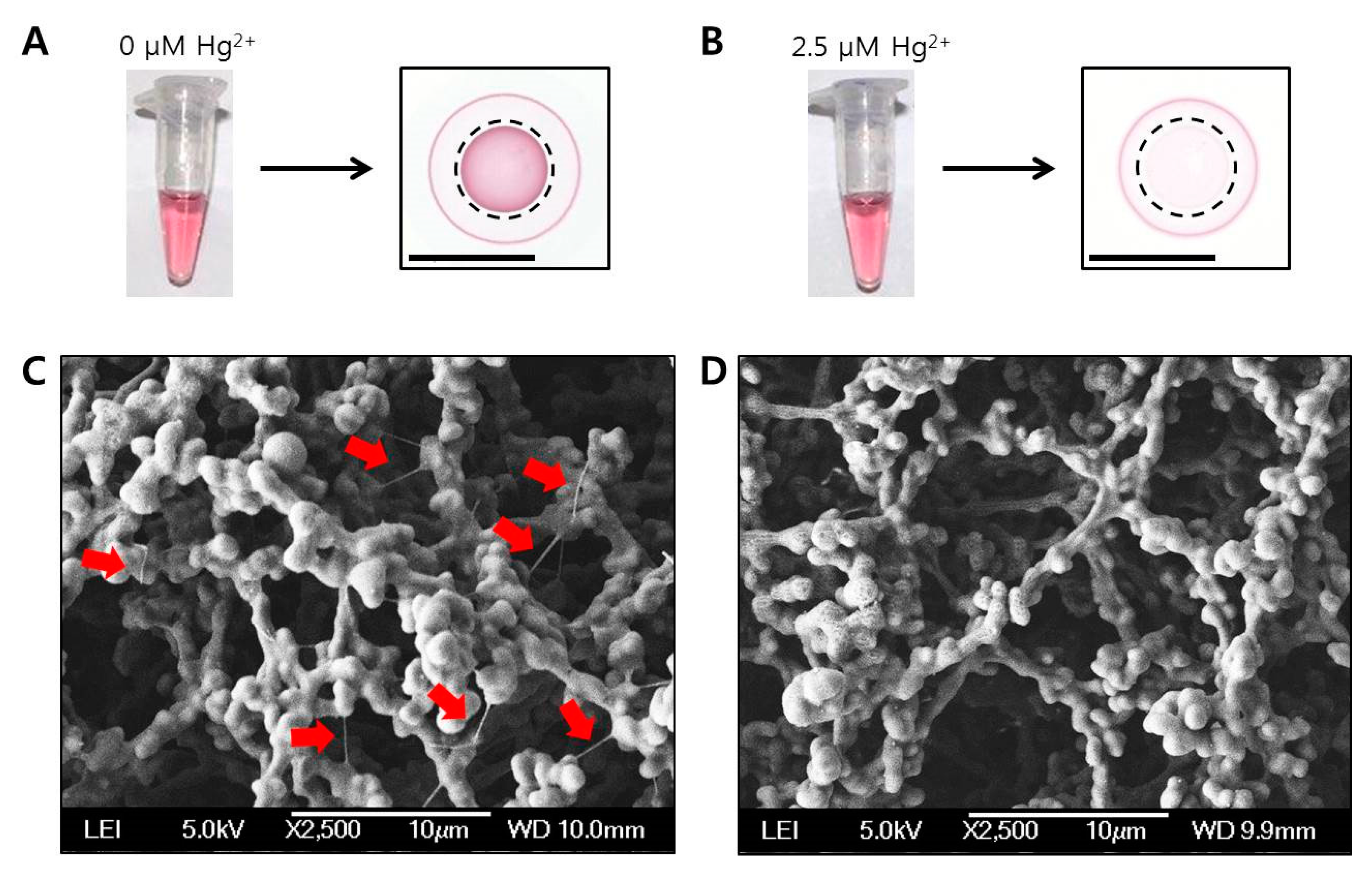
2.4. Different Aspects of Concentric Spot Generation on the Paper According to the Presence of Hg2+
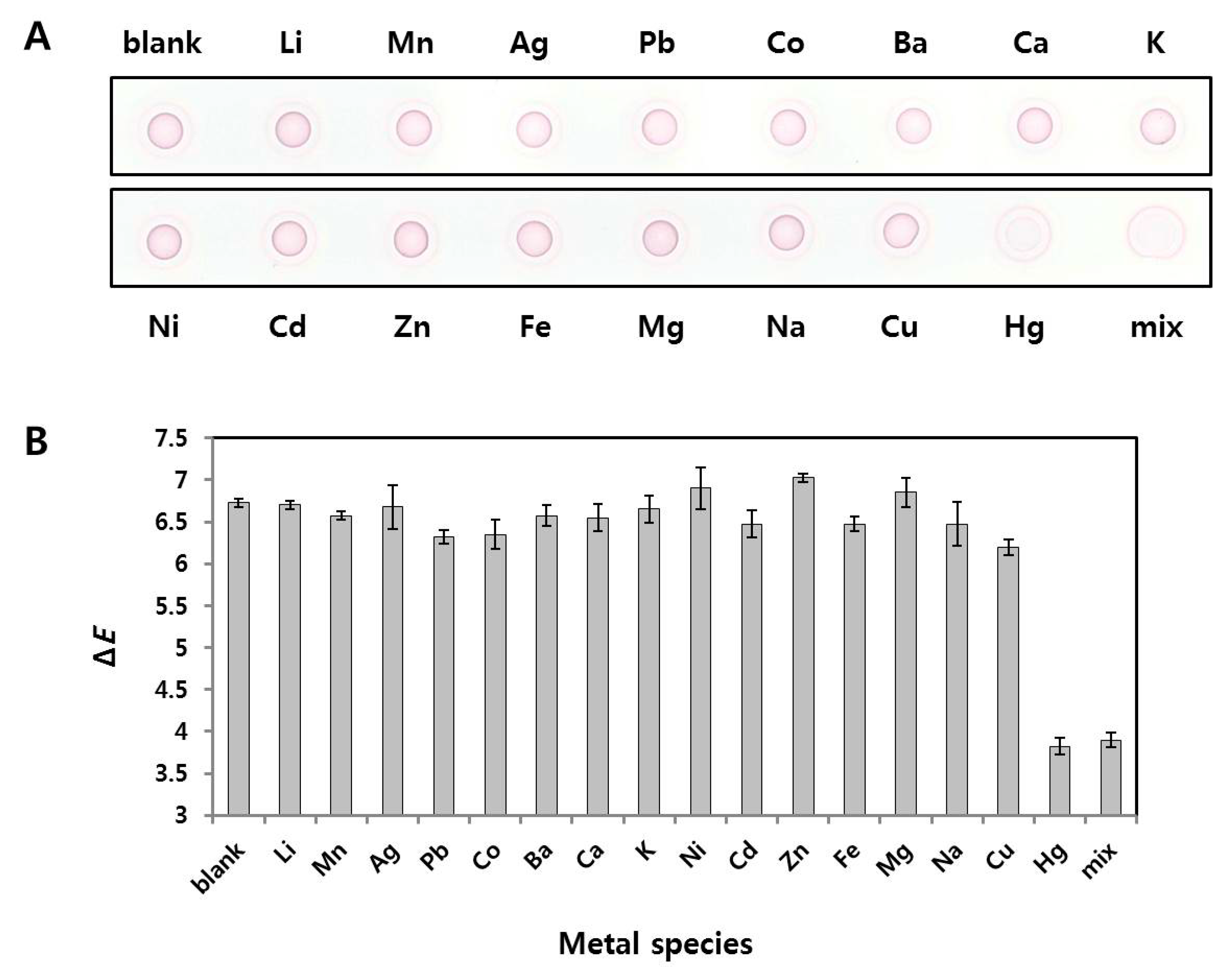
2.5. Selectivity of Radial Flow Assay for Hg2+
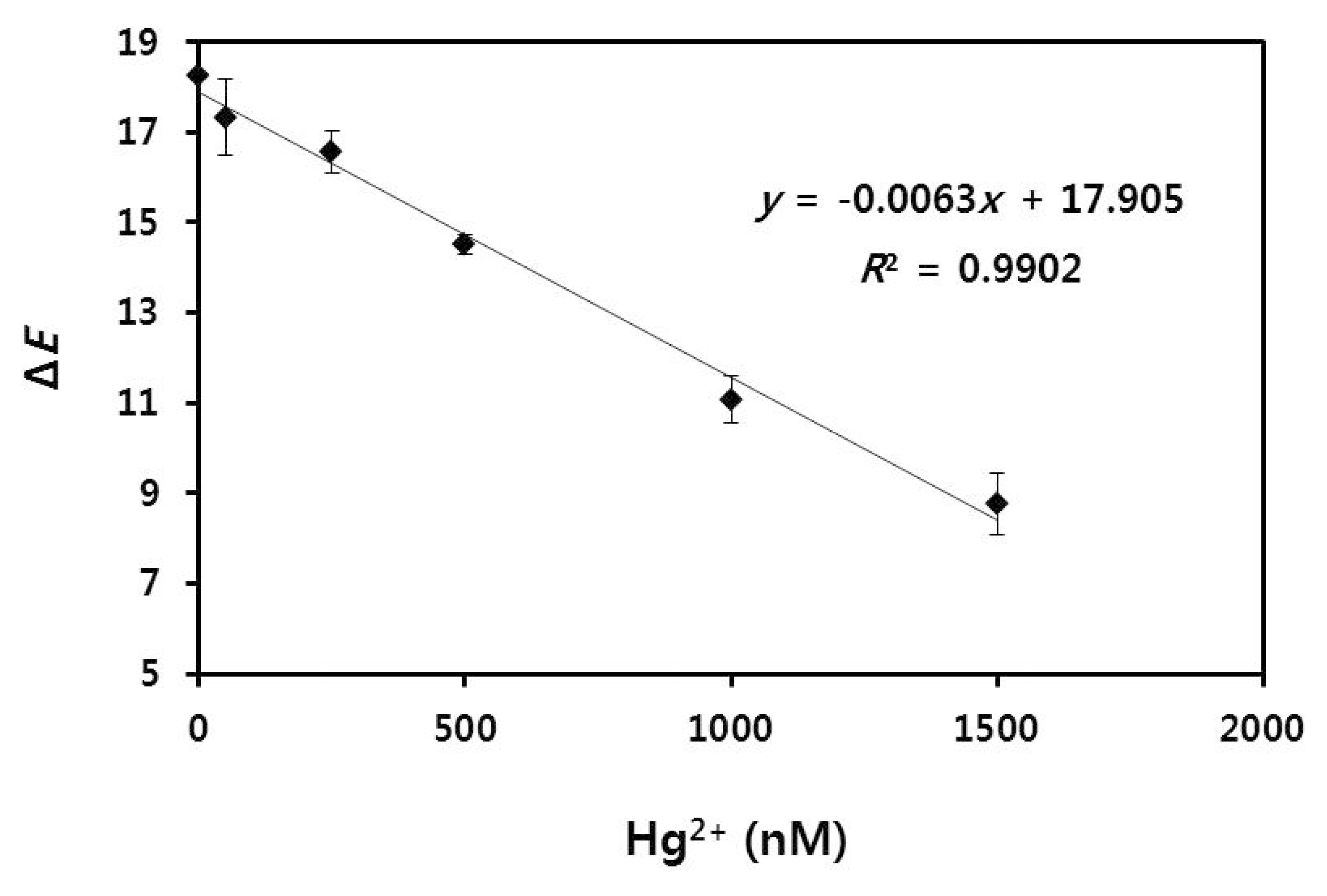
2.6. Quantification of Hg2+ Using Radial Flow Assay
2.7. Analysis of Hg2+ in Tap Water Samples
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Preparations for Circular Template DNA
3.3. Preparations for DNA-Modified AuNPs
3.4. RCA Reaction Test
3.5. Selectivity for Hg2+
3.6. Quantification of Hg2+ by Radial Flow Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baughman, T.A. Elemental mercury spills. Environ. Health Perspect. 2006, 114, 114–147. [Google Scholar] [CrossRef]
- Förstner, U.; Wittmann, G.T.W. Metal Pollution in the Aquatic Environment; Springer: Berlin, Germany, 2012. [Google Scholar]
- Nolan, E.M.; Lippard, S.J. Tools and tactics for the optical detection of mercuric ion. Chem. Rev. 2008, 108, 3443–3480. [Google Scholar] [CrossRef] [PubMed]
- Tchounwou, P.B.; Ayensu, W.K.; Ninashvili, N.; Sutton, D. Environmental exposure to mercury and its toxicopathologic implications for public health. Environ. Toxicol. 2003, 18, 149–175. [Google Scholar] [CrossRef] [PubMed]
- De Smaele, T.; Moens, L.; Dams, R.; Sandra, P.; Van der Eycken, J.; Vandyck, J. Sodium tetra (n-propyl) borate: A novel aqueous in situ derivatization reagent for the simultaneous determination of organomercury, -lead and -tin compounds with capillary gas chromatography–inductively coupled plasma mass spectrometry. J. Chromatogr. A 1998, 793, 99–106. [Google Scholar] [CrossRef]
- Ghaedi, M.; Fathi, M.R.; Shokrollahi, A.; Shajarat, F. Highly selective and sensitive preconcentration of mercury ion and determination by cold vapor atomic absorption spectroscopy. Anal. Lett. 2006, 39, 1171–1185. [Google Scholar] [CrossRef]
- Bache, C.A.; Lisk, D.J. Gas-chromatographic determination of organic mercury compounds by emission spectrometry in a helium plasma. Application to the analysis of methylmercuric salts in fish. Anal. Chem. 1971, 43, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Liu, B.; Liu, F.; Han, M.-Y.; Zhang, Z. Fluorescence “turn on” detection of mercuric ion based on bis (dithiocarbamato) copper (II) complex functionalized carbon nanodots. Anal. Chem. 2014, 86, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- LI, X.; Wu, Y.; Liu, Y.; Zou, X.; Yao, L.; Li, F.; Feng, W. Cyclometallated ruthenium complex-modified upconversion nanophosphors for selective detection of Hg2+ ions in water. Nanoscale 2014, 6, 1020–1028. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, S.; Swierczewska, M.; Huang, X.; Bhirde, A.A.; Sun, J.; Wang, Z.; Yang, M.; Jiang, X.; Chen, X. Highly robust, recyclable displacement assay for mercuric ions in aqueous solutions and living cells. ACS Nano 2012, 6, 10999–11008. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhang, L.; Zhang, S.; Yang, Y.; Chen, X.; Zhang, M. Fluorescent carbon nanoparticles for the fluorescent detection of metal ions. Biosens. Bioelectron. 2015, 63, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cheng, F.; Li, J.; Zhu, J.J.; Lu, Y. Fluorescent nanoprobes for sensing and imaging of metal ions: Recent advances and future perspectives. Nano Today 2016, 11, 309–329. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Brook, M.A.; Li, Y. Design of gold nanoparticle-based colorimetric biosensing assays. ChemBioChem 2008, 9, 2363–2371. [Google Scholar] [CrossRef] [PubMed]
- Chansuvarn, W.; Tuntulani, T.; Imyim, A. Colorimetric detection of mercury (II) based on gold nanoparticles, fluorescent gold nanoclusters and other gold-based nanomaterials. Trends Anal. Chem. 2015, 65, 83–96. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Pal, T. Interparticle coupling effect on the surface plasmon resonance of gold nanoparticles: From theory to applications. Chem. Rev. 2007, 107, 4797–4862. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.L.; Jung, C.; Parab, H.; Li, T.; Park, H.G. Direct colorimetric diagnosis of pathogen infections by utilizing thiol-labeled PCR primers and unmodified gold nanoparticles. Biosens. Bioelectron. 2010, 25, 1941–1946. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Han, M.S.; Mirkin, C.A. Colorimetric detection of mercuric ion (Hg2+) in aqueous media using DNA-functionalized gold nanoparticles. Angew. Chem. Int. Ed. 2007, 46, 4093–4096. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.G.; Zhou, X.; Liu, J.; Wang, R.; Liu, L.; Yu, B.; He, M.; Shia, H. Utilization of unmodified gold nanoparticles for label-free detection of mercury (II): Insight into rational design of mercury-specific oligonucleotides. J. Hazard. Mater. 2017, 321, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.W.; Hsieh, Y.T.; Huang, C.C.; Lin, Z.H.; Chang, H.T. Detection of mercury (II) based on Hg2+–DNA complexes inducing the aggregation of gold nanoparticles. Chem. Commun. 2008, 19, 2242–2244. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Togashi, H.; Tashiro, M.; Yamaguchi, H.; Oda, S.; Kudo, M.; Tanaka, Y.; Kondo, Y.; Sawa, R.; Fujimoto, T.; et al. MercuryII-mediated formation of thymine−HgII−thymine base pairs in DNA duplexes. J. Am. Chem. Soc. 2006, 128, 2172–2173. [Google Scholar] [CrossRef] [PubMed]
- Yamane, T.; Davidson, N. On the complexing of desoxyribonucleic acid (DNA) by mercuric ion1. J. Am. Chem. Soc. 1961, 83, 2599–2607. [Google Scholar] [CrossRef]
- Ono, A.; Togashi, H. Highly selective oligonucleotide-based sensor for mercury (II) in aqueous solutions. Angew. Chem. Int. Ed. 2004, 43, 4300–4302. [Google Scholar] [CrossRef] [PubMed]
- Torabi, S.-F.; Lu, Y. Small-molecule diagnostics based on functional DNA nanotechnology: A dipstick test for mercury. Faraday Discuss. 2011, 149, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Wang, F.; Liu, X. One-step, room temperature, colorimetric detection of mercury (Hg2+) using DNA/nanoparticle conjugates. J. Am. Chem. Soc. 2008, 130, 3244–3245. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.K.; Luppa, P.B.; Yeo, L.Y.; Ozcan, A.; Luong, J.H. Emerging technologies for next-generation point-of-care testing. Trends Biotechnol. 2015, 33, 692–705. [Google Scholar] [CrossRef] [PubMed]
- De Puig, H.; Bosch, I.; Gehrke, L.; Hamad-Schifferli, K. Challenges of the Nano-Bio Interface in Lateral Flow and Dipstick Immunoassays. Trends Biotechnol. 2017, 35, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.G.; DiTucci, M.J.; Phillips, S.T. Quantifying analytes in paper-based microfluidic devices without using external electronic readers. Angew. Chem. Int. Ed. 2012, 51, 12707–12710. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.S.; Tsaloglou, M.N.; Sisley, T.; Christodouleas, D.; Chen, A.; Milette, J.; Whitesides, G.M. Sliding-strip microfluidic device enables ELISA on paper. Biosens. Bioelectron. 2018, 99, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Gritsenko, D.; Feng, S.; Teh, Y.C.; Lu, X.; Xu, J. Detection of heavy metal by paper-based microfluidics. Biosens. Bioelectron. 2016, 83, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, J.C.; DeGregory, P.R.; Crooks, R.M. New functionalities for paper-based sensors lead to simplified user operation, lower limits of detection, and new applications. Annu. Rev. Anal. Chem. 2016, 9, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Sher, M.; Zhuang, R.; Demirci, U.; Asghara, W. Paper-based analytical devices for clinical diagnosis: Recent advances in the fabrication techniques and sensing mechanisms. Expert Rev. Mol. Diagn. 2017, 17, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Fang, X.; Li, H.; Cao, H.; Kong, J. A simple paper-based colorimetric device for rapid mercury (II) assay. Sci. Rep. 2016, 6, 31948. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.H.; Chen, W.Y.; Yen, Y.C.; Wang, C.W.; Chang, H.T.; Chen, C.F. Detection of mercury(II) ions using colorimetric gold nanoparticles on paper-based analytical devices. Anal. Chem. 2014, 86, 6843–6849. [Google Scholar] [CrossRef] [PubMed]
- Han, K.N.; Choi, J.S.; Kwon, J. Gold nanozyme-based paper chip for colorimetric detection of mercury ions. Sci. Rep. 2017, 7, 2806. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ali, M.M.; Brook, M.A.; Li, Y. Rolling circle amplification: Applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. 2008, 47, 6330–6337. [Google Scholar] [CrossRef] [PubMed]
- Banér, J.; Nilsson, M.; Mendel-Hartvig, M.; Landegren, U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998, 26, 5073–5078. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Qiang, S.; Xing, D. An electrochemiluminescent assay for high sensitive detection of mercury (II) based on isothermal rolling circular amplification. Anal. Chim. Acta 2012, 713, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, W.; Yang, Q.; Shi, C.; Cao, L. Cocaine detection via rolling circle amplification of short DNA strand separated by magnetic beads. Biosens. Bioelectron. 2011, 26, 3309–3312. [Google Scholar] [CrossRef] [PubMed]
- Tehrani, S.J.; Aliabadian, M.; Fata, A.; Najafzadeh, M.J. Rolling circle amplification (RCA): An approach for quick detection and identification of fungal species. J. Mycol. Res. 2014, 1, 55–62. [Google Scholar]
- Jung, I.Y.; Lee, E.H.; Suh, A.Y.; Lee, S.J.; Lee, H. Oligonucleotide-based biosensors for in vitro diagnostics and environmental hazard detection. Anal. Bioanal. Chem. 2016, 408, 2383–2406. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Mercury in Drinking Water. Background Document for Development of WHO Guidelines for Drinking-Water Quality; World Health Organization: Geneva, Switzerland, 2005. [Google Scholar]

| Samples No. | Added (nM) | Within-Assay | Between-Assay | ||||
|---|---|---|---|---|---|---|---|
| Found (nM) (Mean a ± SD) | CV b (%) | Recovery (%) | Found (nM) (Mean a ± SD) | CV b (%) | Recovery (%) | ||
| 1 | 150 | 140.5 ± 0.32 | 1.9 | 93.7 | 162.7 ± 0.19 | 1.2 | 108.5 |
| 2 | 450 | 454.8 ± 0.23 | 1.6 | 101.1 | 432.5 ± 0.5 | 3.3 | 96.1 |
| 3 | 700 | 705.6 ± 0.38 | 2.8 | 100.8 | 696.0 ± 0.64 | 4.7 | 99.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.-Y.; Lim, M.-C.; Woo, M.-A.; Jun, B.-H. Radial Flow Assay Using Gold Nanoparticles and Rolling Circle Amplification to Detect Mercuric Ions. Nanomaterials 2018, 8, 81. https://doi.org/10.3390/nano8020081
Kim T-Y, Lim M-C, Woo M-A, Jun B-H. Radial Flow Assay Using Gold Nanoparticles and Rolling Circle Amplification to Detect Mercuric Ions. Nanomaterials. 2018; 8(2):81. https://doi.org/10.3390/nano8020081
Chicago/Turabian StyleKim, Tai-Yong, Min-Cheol Lim, Min-Ah Woo, and Bong-Hyun Jun. 2018. "Radial Flow Assay Using Gold Nanoparticles and Rolling Circle Amplification to Detect Mercuric Ions" Nanomaterials 8, no. 2: 81. https://doi.org/10.3390/nano8020081