Detection of Cardiac Biomarkers Using Single Polyaniline Nanowire-Based Conductometric Biosensors
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Fabrication of Single PANI Nanowire
2.3. Functionalization of Single PANI Nanowire
2.4. Preparation of Microfluidic Channel
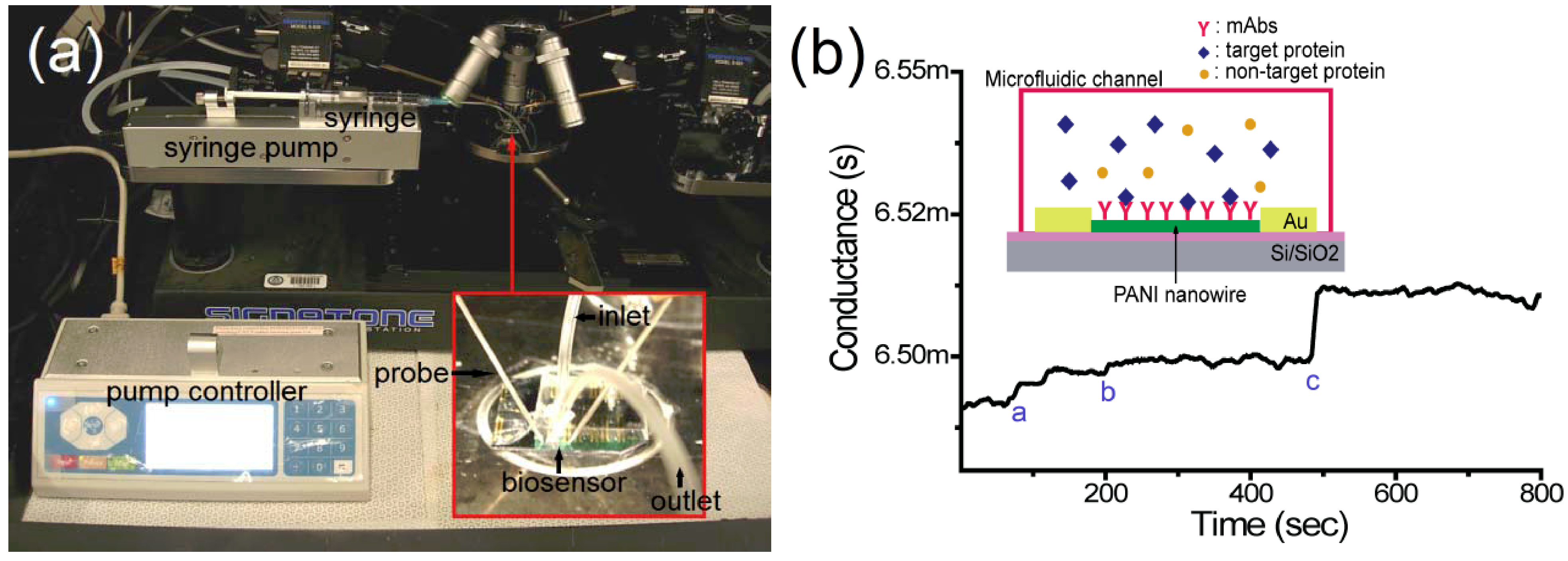
2.5. Detection of Target Proteins on the Nanowire Biosensor
3. Results and Discussion
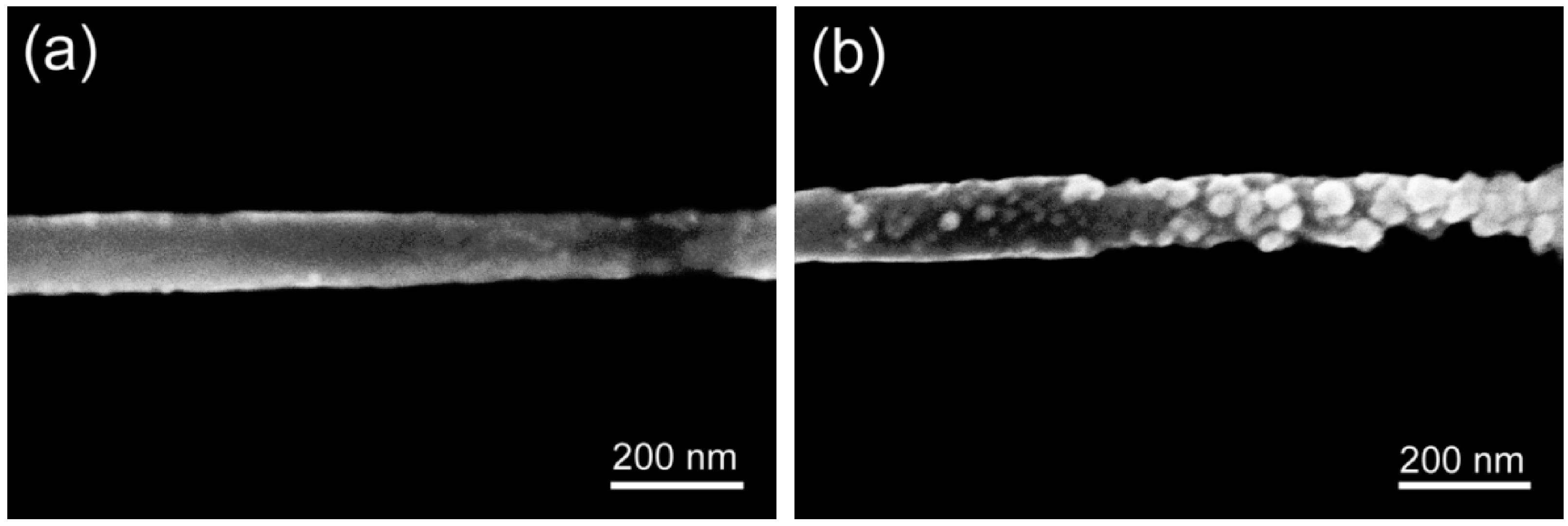
3.1. Functionalization of Single PANI Nanowires with mAbs
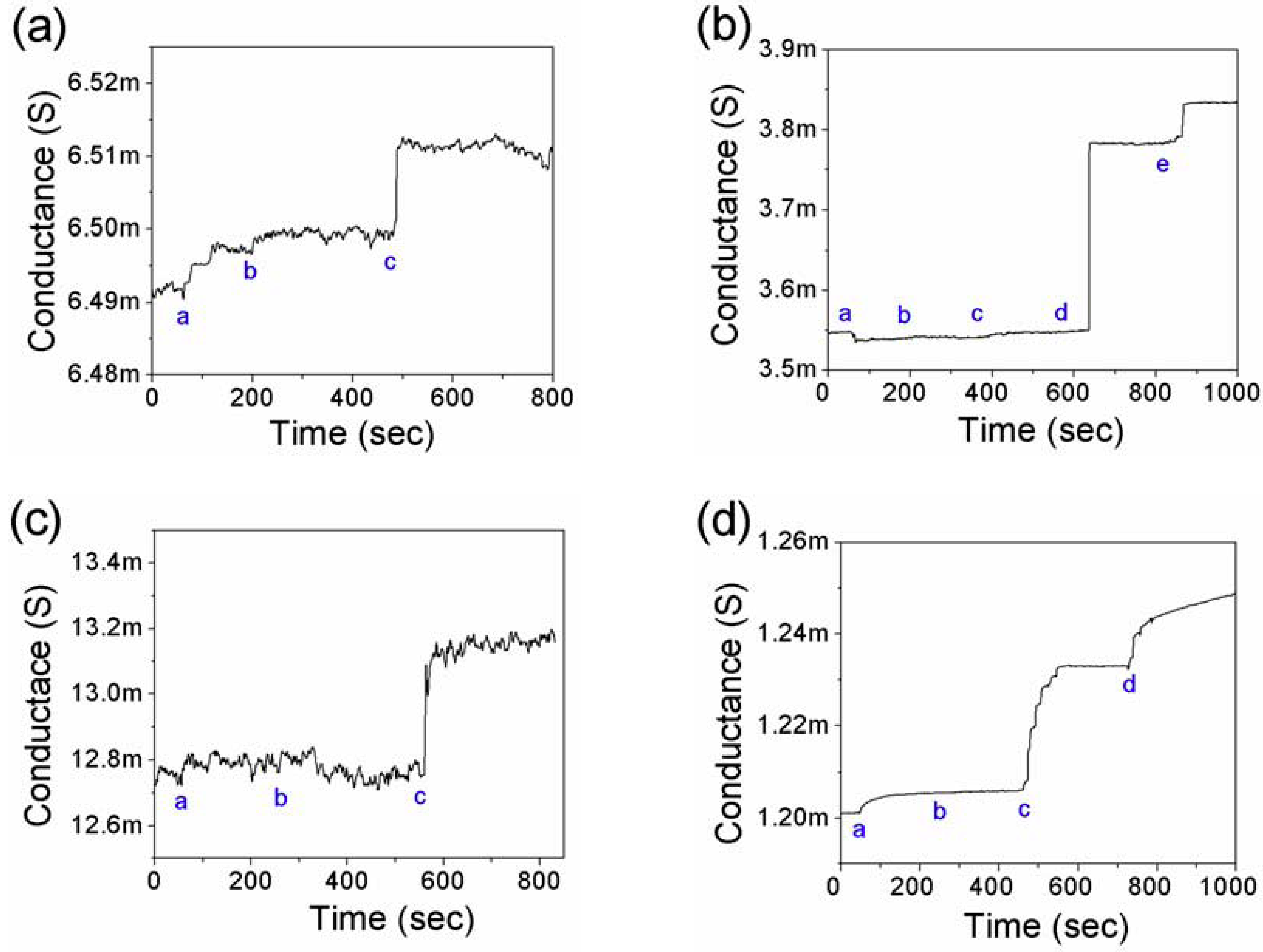
3.2. Detection of Cardiac Biomarkers
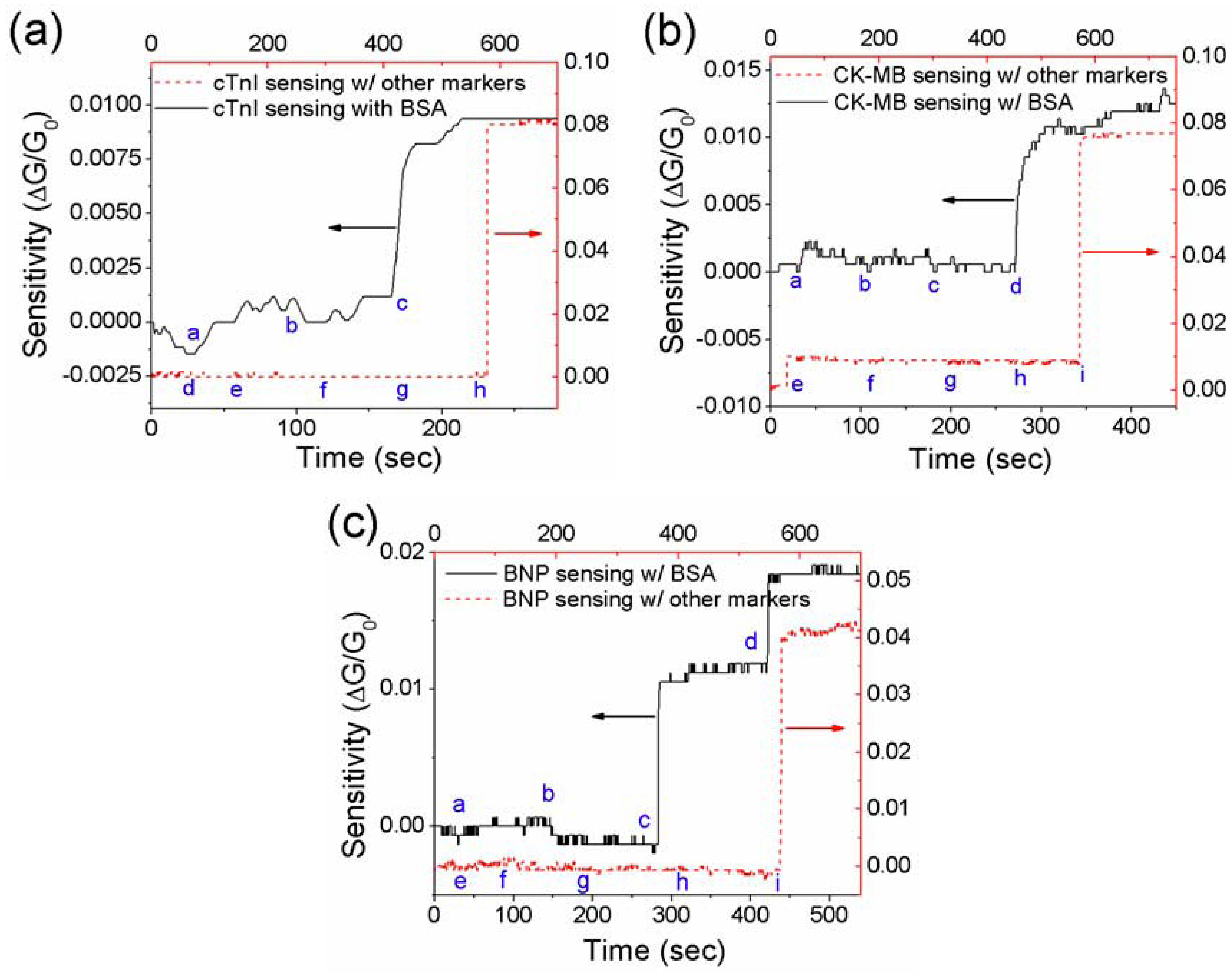
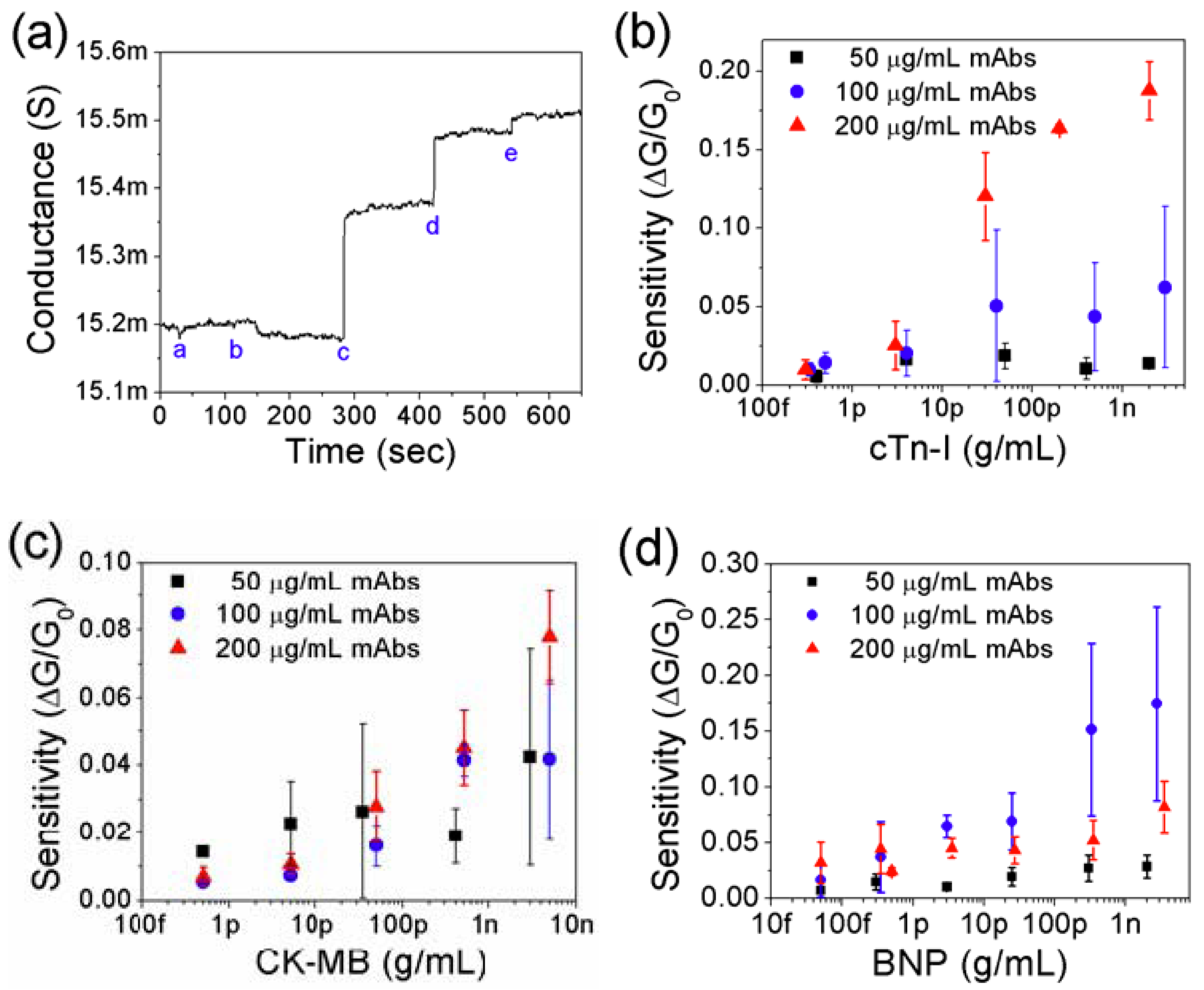
3.3. Effect of Net Surface Charge on the Single PANI Nanowire Biosensor
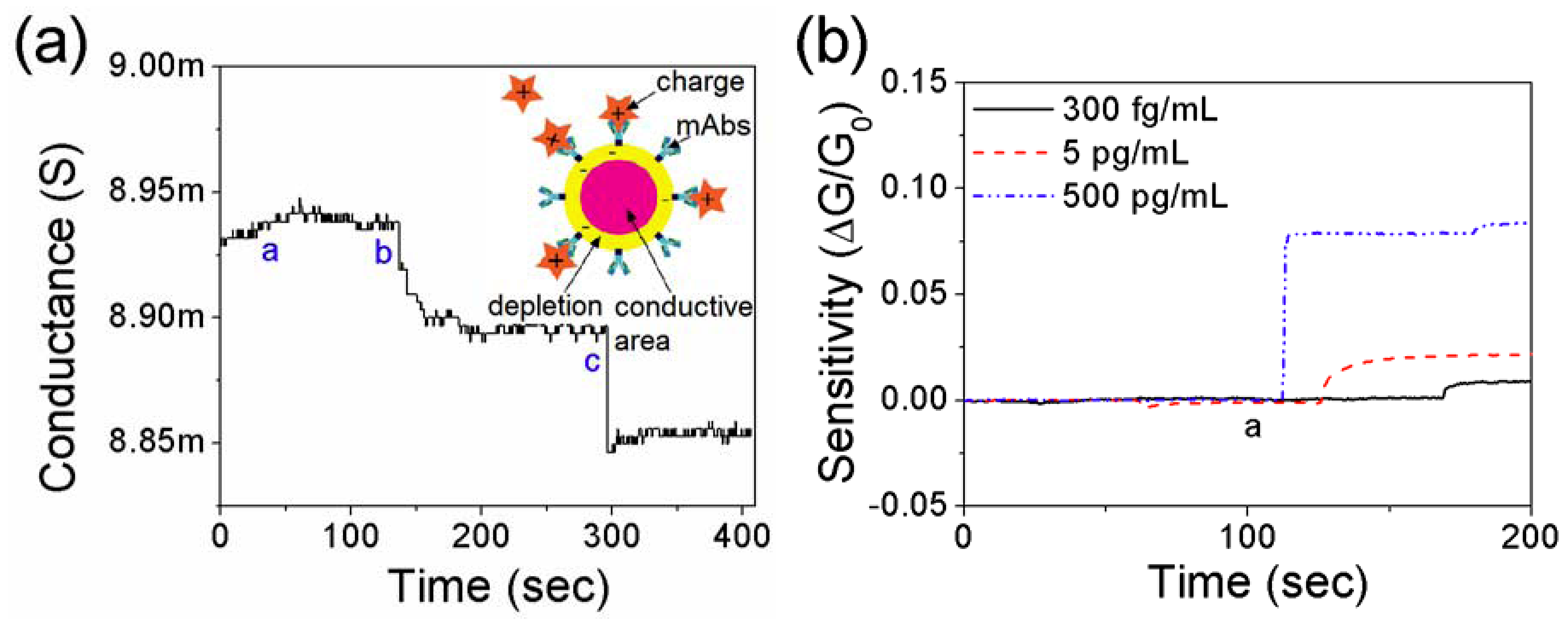
4. Conclusions
Acknowledgement
References
- Tang, W.H.W.; Francis, G.S.; Morrow, D.A.; Newby, L.K.; Cannon, C.P.; Jesse, R.L.; Storrow, A.B.; Christenson, R.H.; Apple, F.S.; Ravkilde, J.; et al. National academy of clinical biochemistry laboratory medicine practice guidelines: Clinical utilization of cardiac biomarker testing in heart failure. Circulation 2007, 116, E99–E109. [Google Scholar]
- Adams, J.E.; Bodor, G.S.; Davilaroman, V.G.; Delmez, J.A.; Apple, F.S.; Ladenson, J.H.; Jaffe, A.S. Cardiac Troponin-I—A marker with high specificity for cardiac injury. Circulation 1993, 88, 101–106. [Google Scholar]
- Etievent, J.-P.; Chocron, S.; Toubin, G.; Taberlet, C.; Alwan, K.; Clement, F.; Cordier, A.; Schipman, N.; Kantelip, J.-P. Use of cardiac troponin I as a marker of perioperative myocardial ischemia. Ann. Thorac. Surg. 1995, 59, 1192–1194. [Google Scholar]
- Apple, F.S.; Jesse, R.L.; Newby, L.K.; Wu, A.H.B.; Christenson, R.H.; Cannon, C.P.; Francis, G.; Jesse, R.; Morrow, D.A.; Ravkilde, J.; et al. National academy of clinical biochemistry and ifcc committee for standardization of markers of cardiac damage laboratory medicine practice guidelines: Analytical issues for biochemical markers of acute coronary syndromes. Clin. Chem. 2007, 53, 547–551. [Google Scholar]
- Masson, J.F.; Obando, L.; Beaudoin, S.; Booksh, K. Sensitive and real-time fiber-optic-based surface plasmon resonance sensors for myoglobin and cardiac troponin I. Talanta 2004, 62, 865–870. [Google Scholar]
- Bhayana, V.; Gougoulias, T.; Cohoe, S.; Henderson, A. Discordance between results for serum troponin T and troponin I in renal disease. Clin. Chem. 1995, 41, 312–317. [Google Scholar]
- Adams, J., 3rd; Abendschein, D.; Jaffe, A. Biochemical markers of myocardial injury. Is MB creatine kinase the choice for the 1990s? Circulation 1993, 88, 750–763. [Google Scholar]
- Caulum, M.M.; Murphy, B.M.; Ramsay, L.M.; Henry, C.S. Detection of cardiac biomarkers using micellar electrokinetic chromatography and a cleavable tag immunoassay. Anal. Chem. 2007, 79, 5249–5256. [Google Scholar]
- Darain, F.; Yager, P.; Gan, K.L.; Tjin, S.C. On-chip detection of myoglobin based on fluorescence. Biosens. Bioelectron. 2009, 24, 1744–1750. [Google Scholar]
- Masson, J.-F.; Battaglia, T.M.; Khairallah, P.; Beaudoin, S.; Booksh, K.S. Quantitative measurement of cardiac markers in undiluted serum. Anal. Chem. 2006, 79, 612–619. [Google Scholar]
- Kurita, R.; Yokota, Y.; Sato, Y.; Mizutani, F.; Niwa, O. On-chip enzyme immunoassay of a cardiac marker using a microfluidic device combined with a portable surface plasmon resonance system. Anal. Chem. 2006, 78, 5525–5531. [Google Scholar]
- Chua, J.H.; Chee, R.-E.; Agarwal, A.; Wong, S.M.; Zhang, G.-J. Label-free electrical detection of cardiac biomarker with complementary metal-oxide semiconductor-compatible silicon nanowire sensor arrays. Anal. Chem. 2009, 81, 6266–6271. [Google Scholar]
- Lin, T.-W.; Hsieh, P.-J.; Lin, C.-L.; Fang, Y.-Y.; Yang, J.-X.; Tsai, C.-C.; Chiang, P.-L.; Pan, C.-Y.; Chen, Y.-T. Label-free detection of protein-protein interactions using a calmodulin-modified nanowire transistor. Proc. Natl. Acad..Sci. USA 2010, 107, 1047–1052. [Google Scholar]
- Plowman, T.E.; Durstchi, J.D.; Wang, H.K.; Christensen, D.A.; Herron, J.N.; Reichert, W.M. Multiple-analyte fluoroimmunoassay using an integrated optical waveguide sensor. Anal. Chem. 1999, 71, 4344–4352. [Google Scholar]
- Dutra, R.F.; Kubota, L.T. An SPR immunosensor for human cardiac troponin T using specific binding avidin to biotin at carboxymethyldextran-modified gold chip. Clin. Chim. Acta 2007, 376, 114–120. [Google Scholar]
- Allen, B.L.; Kichambare, P.D.; Star, A. Carbon nanotube field-effect-transistor-based biosensors. Adv. Mater. 2007, 19, 1439–1451. [Google Scholar]
- Balasubramanian, K. Challenges in the use of 1D nanostructures for on-chip biosensing and diagnostics: A review. Biosens. Bioelectron. 2010, 26, 1195–1204. [Google Scholar]
- Curreli, M.; Rui, Z.; Ishikawa, F.N.; Hsiao-Kang, C.; Cote, R.J.; Chongwu, Z.; Thompson, M.E. Real-time, label-free detection of biological entities using nanowire-based FETs. IEEE Trans. Nanotechnol. 2008, 7, 651–667. [Google Scholar]
- Wang, J. Nanomaterial-based electrochemical biosensors. Analyst 2005, 130, 421–426. [Google Scholar]
- Gooding, J.J. Nanoscale biosensors: Significant advantages over larger devices? Small 2006, 2, 313–315. [Google Scholar]
- Bockrath, M.; Markovic, N.; Shepard, A.; Tinkham, M.; Gurevich, L.; Kouwenhoven, L.P.; Wu, M.W.; Sohn, L.L. Scanned conductance microscopy of carbon nanotubes and λ-DNA. Nano Lett. 2002, 2, 187–190. [Google Scholar]
- Ishikawa, F.N.; Chang, H.-K.; Curreli, M.; Liao, H.-I.; Olson, C.A.; Chen, P.-C.; Zhang, R.; Roberts, R.W.; Sun, R.; Cote, R.J.; et al. Label-free, electrical detection of the SARS virus n-protein with nanowire biosensors utilizing antibody mimics as capture probes. ACS Nano 2009, 3, 1219–1224. [Google Scholar]
- Gao, Z.; Agarwal, A.; Trigg, A.D.; Singh, N.; Fang, C.; Tung, C.-H.; Fan, Y.; Buddharaju, K.D.; Kong, J. Silicon nanowire arrays for label-free detection of DNA. Anal. Chem. 2007, 79, 3291–3297. [Google Scholar]
- Malhotra, B.D.; Chaubey, A.; Singh, S.P. Prospects of conducting polymers in biosensors. Anal. Chim. Acta 2006, 578, 59–74. [Google Scholar]
- Heeger, P.S.; Heeger, A.J. Making sense of polymer-based biosensors. Proc. Natl. Acad. Sci. USA 1999, 96, 12219–12221. [Google Scholar]
- Tolani, S.; Craig, M.; DeLong, R.; Ghosh, K.; Wanekaya, A. Towards biosensors based on conducting polymer nanowires. Anal. Bioanal. Chem. 2009, 393, 1225–1231. [Google Scholar]
- Gerard, M.; Chaubey, A.; Malhotra, B.D. Application of conducting polymers to biosensors. Biosens. Bioelectron. 2002, 17, 345–359. [Google Scholar]
- Adhikari, B.; Majumdar, S. Polymers in sensor applications. Prog. Polym. Sci. 2004, 29, 699–766. [Google Scholar]
- Ahuja, T.; Mir, I.A.; Kumar, D. Rajesh biomolecular immobilization on conducting polymers for biosensing applications. Biomaterials 2007, 28, 791–805. [Google Scholar]
- Briseno, A.L.; Mannsfeld, S.C.B.; Jenekhe, S.A.; Bao, Z.; Xia, Y. Introducing organic nanowire transistors. Mater. Today 2008, 11, 38–47. [Google Scholar]
- He, H.; Zhu, J.; Tao, N.J.; Nagahara, L.A.; Amlani, I.; Tsui, R. A conducting polymer nanojunction Switch. J. Am. Chem. Soc. 2001, 123, 7730–7731. [Google Scholar]
- Arter, J.A.; Taggart, D.K.; McIntire, T.M.; Penner, R.M.; Weiss, G.A. Virus-PEDOT nanowires for biosensing. Nano Lett. 2010, 10, 4858–4862. [Google Scholar]
- Bangar, M.A.; Shirale, D.J.; Chen, W.; Myung, N.V.; Mulchandani, A. Single conducting polymer nanowire chemiresistive label-free immunosensor for cancer biomarker. Anal. Chem. 2009, 81, 2168–2175. [Google Scholar]
- Yoon, H.; Lee, S.H.; Kwon, O.S.; Song, H.S.; Oh, E.H.; Park, T.H.; Jang, J. Polypyrrole nanotubes conjugated with human olfactory receptors: High-performance transducers for fet-type bioelectronic noses. Angew. Chem. Int. Ed. 2009, 48, 2755–2758. [Google Scholar]
- Yun, M.; Myung, N.V.; Vasquez, R.P.; Lee, C.; Menke, E.; Penner, R.M. Electrochemically grown wires for individually addressable sensor arrays. Nano Lett. 2004, 4, 419–422. [Google Scholar]
- Lee, I.; Il Park, H.; Park, S.; Kim, M.J.; Yun, M. Highly reproducible single polyaniline nanowire using electrophoresis method. Nano 2008, 3, 75–82. [Google Scholar]
- Duffy, D.C.; McDonald, J.C.; Schueller, O.J.A.; Whitesides, G.M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 1998, 70, 4974–4984. [Google Scholar]
- Nair, P.R.; Alam, M.A. Design considerations of silicon nanowire biosensors. IEEE Trans. Electron. Dev. 2007, 54, 3400–3408. [Google Scholar]
- Ban, N.; Escobar, C.; Garcia, R.; Hasel, K.; Day, J.; Greenwood, A.; McPherson, A. Crystal structure of an idiotype-anti-idiotype Fab complex. Proc. Natl. Acad. Sci. USA 1994, 91, 1604–1608. [Google Scholar]
- Murphy, R.M.; Slayter, H.; Schurtenberger, P.; Chamberlin, R.A.; Colton, C.K.; Yarmush, M.L. Size and structure of antigen-antibody complexes. Electron microscopy and light scattering studies. Biophys. J. 1988, 54, 45–56. [Google Scholar]
- Kwon, O.S.; Park, S.J.; Jang, J. A high-performance VEGF aptamer functionalized polypyrrole nanotube biosensor. Biomaterials 2010, 31, 4740–4747. [Google Scholar]
- Hsiao, C.-Y.; Lin, C.-H.; Hung, C.-H.; Su, C.-J.; Lo, Y.-R.; Lee, C.-C.; Lin, H.-C.; Ko, F.-H.; Huang, T.-Y.; Yang, Y.-S. Novel poly-silicon nanowire field effect transistor for biosensing application. Biosens. Bioelectron. 2009, 24, 1223–1229. [Google Scholar]
- Prina-Mello, A.; Whelan, A.M.; Atzberger, A.; McCarthy, J.E.; Byrne, F.; Davies, G.-L.; Coey, J.M.D.; Volkov, Y.; Gun’ko, Y.K. Comparative flow cytometric analysis of immunofunctionalized nanowire and nanoparticle signatures. Small 2010, 6, 247–255. [Google Scholar]
- Katz, E.; Willner, I. Integrated Nanoparticle–Biomolecule Hybrid systems: Synthesis, properties, and applications. Angew. Chem. Int. Ed. 2004, 43, 6042–6108. [Google Scholar]
- Lee, I.; Luo, X.; Cui, X.T.; Yun, M. Highly sensitive single polyaniline nanowire biosensor for the detection of immunoglobulin G and myoglobin. Biosens. Bioelectron. 2011, 26, 3297–3302. [Google Scholar]
- Luo, X.; Lee, I.; Huang, J.; Yun, M.; Cui, X.T. Ultrasensitive protein detection using an aptamer-functionalized single polyaniline nanowire. Chem. Commun. 2011, 47, 6368–6370. [Google Scholar]
- Huang, S.; Yang, H.; Lakshmanan, R.S.; Johnson, M.L.; Wan, J.; Chen, I.H.; Wikle Iii, H.C.; Petrenko, V.A.; Barbaree, J.M.; Chin, B.A. Sequential detection of salmonella typhimurium and bacillus anthracis spores using magnetoelastic biosensors. Biosens. Bioelectron. 2009, 24, 1730–1736. [Google Scholar]
- Mire-Sluis, A.R.; Barrett, Y.C.; Devanarayan, V.; Koren, E.; Liu, H.; Maia, M.; Parish, T.; Scott, G.; Shankar, G.; Shores, E.; et al. Recommendations for the design and optimization of immunoassays used in the detection of host antibodies against biotechnology products. J. Immunol. Methods 2004, 289, 1–16. [Google Scholar]
- Maraldo, D.; Mutharasan, R. Optimization of antibody immobilization for sensing using piezoelectrically excited-millimeter-sized cantilever (PEMC) sensors. Sens. Actuat. B Chem. 2007, 123, 474–479. [Google Scholar]
- Nyamsi Hendji, A.M.; Jaffrezic-Renault, N.; Martelet, C.; Shul’ga, A.A.; Dzydevich, S.V.; Soldatkin, A.P.; El’skaya, A.V. Enzyme biosensor based on a micromachined interdigitated conductometric transducer: Application to the detection of urea, glucose, acetyl- andbutyrylcholine chlordes. Sens. Actuat. B Chem. 1994, 21, 123–129. [Google Scholar]
- Tang, J.; Huang, J.; Su, B.; Chen, H.; Tang, D. Sandwich-type conductometric immunoassay of alpha-fetoprotein in human serum using carbon nanoparticles as labels. Biochem. Eng. J. 2011, 53, 223–228. [Google Scholar]
- Mizel, S.; Mizel, D. Purification to apparent homogeneity of murine interleukin 1. J. Immunol. 1981, 126, 834–837. [Google Scholar]
- Peronnet, E.; Becquart, L.; Martinez, J.; Charrier, J.-P.; Jolivet-Reynaud, C. Isoelectric point determination of cardiac troponin I forms present in plasma from patients with myocardial infarction. Clin. Chim. Acta 2007, 377, 243–247. [Google Scholar]
- Wevers, R.A.; Wolters, R.J.; Soons, J.B.J. Isoelectric focusing and hybridisation experiments on creatine kinase (EC 2.7.3.2). Clin. Chim. Acta 1977, 78, 271–276. [Google Scholar]
- Rowe, C.A.; Tender, L.M.; Feldstein, M.J.; Golden, J.P.; Scruggs, S.B.; MacCraith, B.D.; Cras, J.J.; Ligler, F.S. Array biosensor for simultaneous identification of bacterial, viral, and protein analytes. Anal. Chem. 1999, 71, 3846–3852. [Google Scholar]
- Elfström, N.; Juhasz, R.; Sychugov, I.; Engfeldt, T.; Karlström, A.E.; Linnros, J. Surface charge sensitivity of silicon nanowires: Size dependence. Nano Lett. 2007, 7, 2608–2612. [Google Scholar]
- Rini, J.; Schulze-Gahmen, U.; Wilson, I. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science 1992, 255, 959–965. [Google Scholar]
- Sheriff, S.; Silverton, E.W.; Padlan, E.A.; Cohen, G.H.; Smith-Gill, S.J.; Finzel, B.C.; Davies, D.R. Three-dimensional structure of an antibody-antigen complex. Proc. Natl. Acad. Sci. USA 1987, 84, 8075–8079. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lee, I.; Luo, X.; Huang, J.; Cui, X.T.; Yun, M. Detection of Cardiac Biomarkers Using Single Polyaniline Nanowire-Based Conductometric Biosensors. Biosensors 2012, 2, 205-220. https://doi.org/10.3390/bios2020205
Lee I, Luo X, Huang J, Cui XT, Yun M. Detection of Cardiac Biomarkers Using Single Polyaniline Nanowire-Based Conductometric Biosensors. Biosensors. 2012; 2(2):205-220. https://doi.org/10.3390/bios2020205
Chicago/Turabian StyleLee, Innam, Xiliang Luo, Jiyong Huang, Xinyan Tracy Cui, and Minhee Yun. 2012. "Detection of Cardiac Biomarkers Using Single Polyaniline Nanowire-Based Conductometric Biosensors" Biosensors 2, no. 2: 205-220. https://doi.org/10.3390/bios2020205



