Multidrug Efflux Systems in Helicobacter cinaedi
Abstract
:1. Introduction
2. Multidrug Efflux Systems in Bacteria
3. Antibacterial Resistance Revealed by the Complete Genome of H. cinaedi
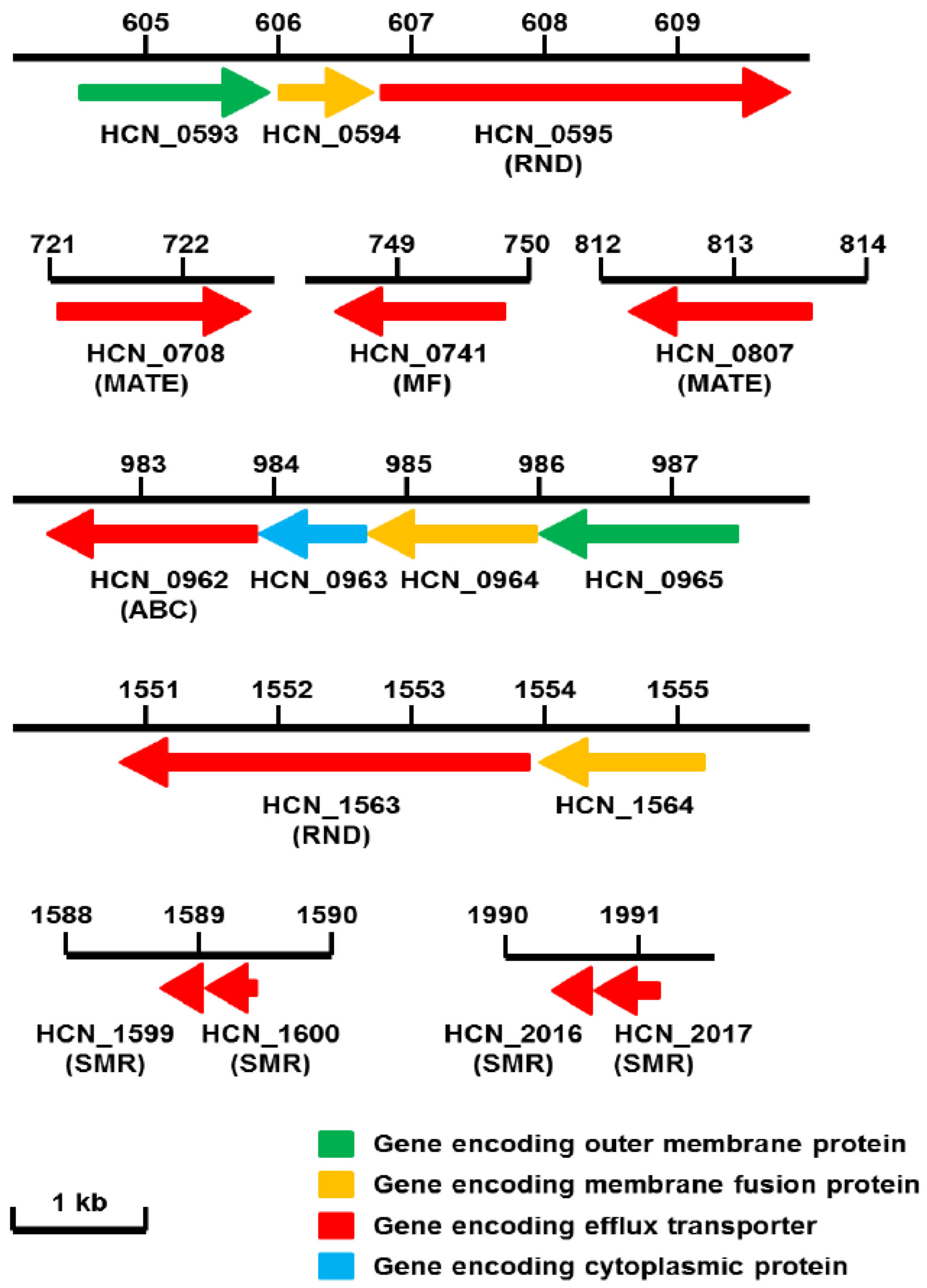
4. RND Efflux Gene Operons of H. cinaedi
5. Structure of the RND Components (HCN_0595 and HCN_1563) of H. cinaedi
| Putative drug transporter | Family | Homologue (Identity (%) (Positives (%))) | ||
|---|---|---|---|---|
| H. hepaticus | H. pylori | C. jejuni | ||
| ATCC 51449 | 26695 | NCTC 11168 | ||
| HCN_0595 | RND | HH0222 (86 (94)) | HefC (58 (78)) | CmeF (38 (59)) |
| HCN_0708 | MATE | HH0167 (81 (90)) | HP1184 (48 (73) | Cj0560 (29 (49)) |
| HCN_0741 | MF | HH1614 (80 (90)) | HP1181 (47 (64)) | CmeG (43 (64)) |
| HCN_0807 | MATE | HH0031 (76 (87)) | HP0759 (40 (62)) | |
| HCN_0962 | ABC | HH1856 (87 (93)) | Cj0607 (32 (57)) | |
| HCN_1563 | RND | HH0174 (88 (95)) | CmeB (53 (73)) | |
| HCN_1599 | SMR | HH0508 (59 (74)) | Cj1174 (54 (76)) | |
| HCN_1600 | SMR | HH0509 (61 (73)) | Cj1173 (40 (64)) | |
| HCN_2016 | SMR | HH1452 (61 (73)) | Cj0309c (57 (73)) | |
| HCN_2017 | SMR | HH1451 (56 (69)) | Cj0309c (57 (73)) | |

6. A Possible Regulator Gene of Multidrug Efflux Systems in H. cinaedi
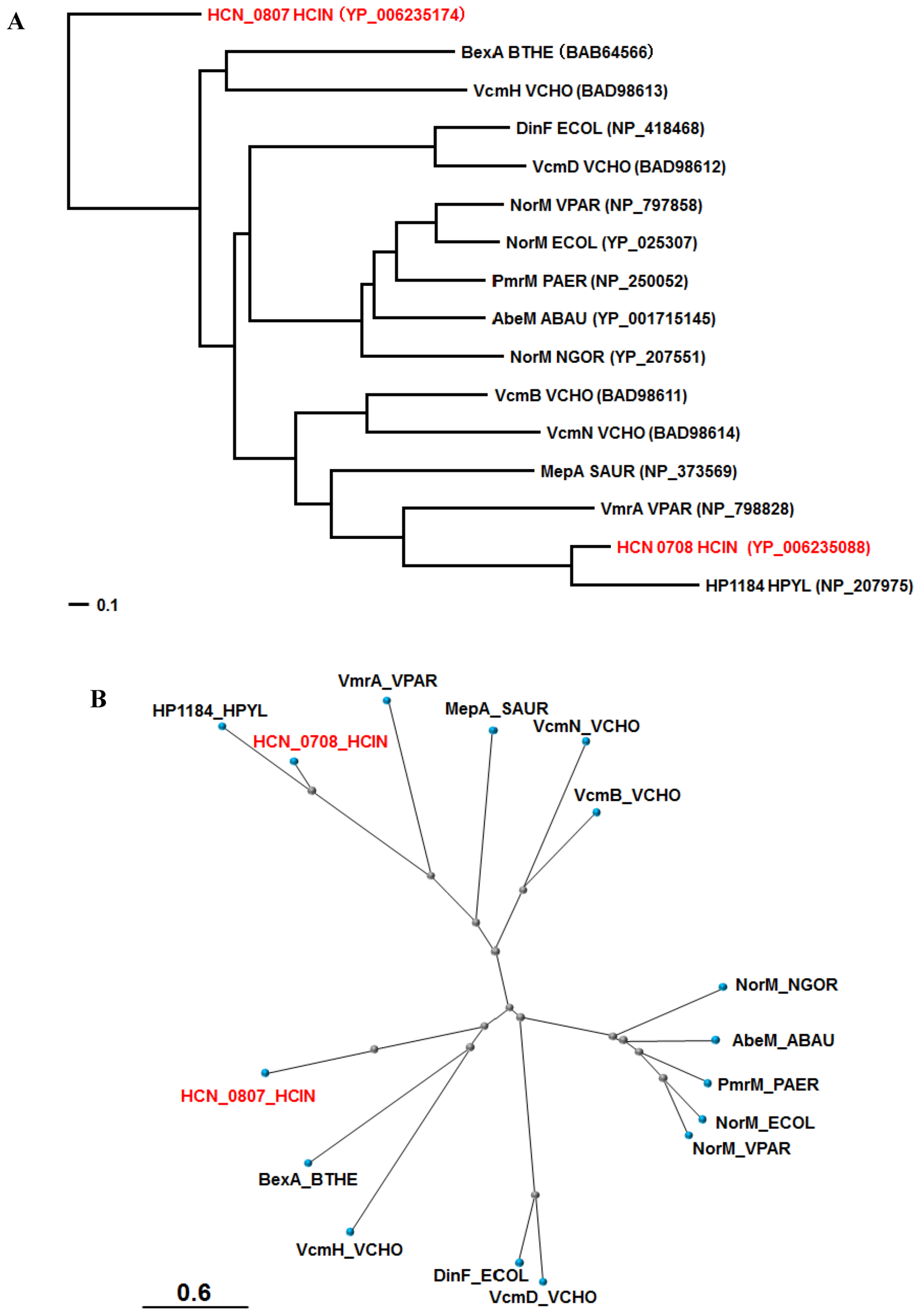
7. C. jejuni CmeG Homologue Identified in H. cinaedi
8. Other Probable Drug Efflux Systems in H. cinaedi
9. Future Perspective
Acknowledgments
Conflict of Interest
References
- Lawson, A.J. Helicobacter. In Manual of Clinical Microbiology, 10th; Versalovic, J., Carroll, K.C., Funke, G., Jorgensen, J.H., Landry, M.L., Warnock, D.W., Eds.; ASM Press: Washington D.C., USA, 2011; Volume 1, pp. 900–915. [Google Scholar]
- Uckay, I.; Garbino, J.; Dietrich, P.Y.; Ninet, B.; Rohner, P.; Jacomo, V. Recurrent Bacteremia with Helicobacter cinaedi: Case Report and Review of the Literature. BMC Infect. Dis. 2006, 6, e86. [Google Scholar] [CrossRef]
- Kitamura, T.; Kawamura, Y.; Ohkusu, K.; Masaki, T.; Iwashita, H.; Sawa, T.; Fujii, S.; Okamoto, T.; Akaike, T. Helicobacter cinaedi Cellulitis and Bacteremia in Immunocompetent Hosts after Orthopedic Surgery. J. Clin. Microbiol. 2007, 45, 31–38. [Google Scholar] [CrossRef]
- Khan, S.; Okamoto, T.; Enomoto, K.; Sakashita, N.; Oyama, K.; Fujii, S.; Sawa, T.; Takeya, M.; Ogawa, H.; Yamabe, H.; et al. Potential Association of Helicobacter cinaedi with Atrial Arrhythmias and Atherosclerosis. Microbiol. Immunol. 2012, 56, 145–154. [Google Scholar]
- Tomida, J.; Kashida, M.; Oinishi, K.; Endo, R.; Morita, Y.; Kawamura, Y. Evaluation of Various Media for Rapid Detection of Helicobacter spp. J. Jpn. Soc. Clin. Microbiol. 2008, 18, 227–235. [Google Scholar]
- Oyama, K.; Khan, S.; Okamoto, T.; Fujii, S.; Ono, K.; Matsunaga, T.; Yoshitake, J.; Sawa, T.; Tomida, J.; Kawamura, Y.; et al. Identification of and Screening for Human Helicobacter cinaedi Infections and Carriers via Nested PCR. J. Clin. Microbiol. 2012, 50, 3893–3900. [Google Scholar] [CrossRef]
- Rimbara, E.; Mori, S.; Matsui, M.; Suzuki, S.; Wachino, J.; Kawamura, Y.; Shen, Z.; Fox, J.G.; Shibayama, K. Molecular Epidemiologic Analysis and Antimicrobial Resistance of Helicobacter cinaedi Isolated from Seven Hospitals in Japan. J. Clin. Microbiol. 2012, 50, 2553–2560. [Google Scholar]
- Tomida, J.; Morita, Y.; Kawamura, Y. Antimicrobial Susceptibility Tests and Resistant Mechanisms of Helicobacter cinaedi. Jan. J. Bacteriol. 2012, 67, 127. [Google Scholar]
- Belzer, C.; Stoof, J.; Breijer, S.; Kusters, J.G.; Kuipers, E.J.; van Vliet, A.H. The Helicobacter hepaticus hefA Gene is Involved in Resistance to Amoxicillin. Helicobacter 2009, 14, 72–79. [Google Scholar] [CrossRef]
- Gibreel, A.; Wetsch, N.M.; Taylor, D.E. Contribution of the CmeABC Efflux Pump to Macrolide and Tetracycline Resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 2007, 51, 3212–3216. [Google Scholar] [CrossRef]
- Morita, Y.; Tomida, J.; Kawamura, Y. Primary Mechanisms Mediating Aminoglycoside Resistance in the Multidrug-resistant Pseudomonas aeruginosa Clinical Isolate PA7. Microbiology 2012, 158, 1071–1083. [Google Scholar] [CrossRef]
- Nikaido, H. Prevention of Drug Access to Bacterial Targets: Permeability Barriers and Active Rfflux. Science 1994, 264, 382–388. [Google Scholar]
- Li, X.Z.; Nikaido, H. Efflux-mediated Drug Resistance in Bacteria: An Update. Drugs 2009, 69, 1555–1623. [Google Scholar]
- Poole, K. Efflux-mediated Antimicrobial Resistance. In Antibiotic Discovery and Development; Dougherty, T.J., Pucci, M.J., Eds.; Springer: New York, NY, USA, 2012; Volume 1, pp. 349–395. [Google Scholar]
- Zgurskaya, H.I.; Nikaido, H. Bypassing the Periplasm: Reconstitution of the AcrAB Multidrug Efflux Pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 1999, 96, 7190–7195. [Google Scholar] [CrossRef]
- Mine, T.; Morita, Y.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. Evidence for Chloramphenicol/H+ Antiport in Cmr (MdfA) System of Escherichia coli and Properties of the Antiporter. J. Biochem. 1998, 124, 187–193. [Google Scholar] [CrossRef]
- Yerushalmi, H.; Lebendiker, M.; Schuldiner, S. EmrE, an Escherichia coli 12-kDa Multidrug Transporter, Exchanges Toxic Cations and H+ and is Soluble in Organic Solvents. J. Biol. Chem. 1995, 270, 6856–6863. [Google Scholar]
- Morita, Y.; Kataoka, A.; Shiota, S.; Mizushima, T.; Tsuchiya, T. NorM of Vibrio parahaemolyticus is an Na(+)-driven Multidrug Efflux Pump. J. Bacteriol. 2000, 182, 6694–6697. [Google Scholar] [CrossRef]
- Kobayashi, N.; Nishino, K.; Yamaguchi, A. Novel Macrolide-specific ABC-type Efflux Transporter in Escherichia coli. J. Bacteriol. 2001, 183, 5639–5644. [Google Scholar]
- Huda, N.; Lee, E.W.; Chen, J.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Molecular Cloning and Characterization of an ABC Multidrug Efflux Pump, VcaM, in Non-O1 Vibrio cholerae. Antimicrob. Agents Chemother. 2003, 47, 2413–2417. [Google Scholar] [CrossRef]
- Mine, T.; Morita, Y.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. Expression in Escherichia coli of a New Multidrug Efflux Pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1999, 43, 415–417. [Google Scholar]
- Goto, T.; Ogura, Y.; Hirakawa, H.; Tomida, J.; Morita, Y.; Akaike, T.; Hayashi, T.; Kawamura, Y. Complete Genome Sequence of Helicobacter cinaedi Strain PAGU 611, Isolated in a Case of Human Bacteremia. J. Bacteriol. 2012, 194, 3744–3745. [Google Scholar] [CrossRef]
- Miyoshi-Akiyama, T.; Takeshita, N.; Ohmagari, N.; Kirikae, T. Complete Genome Sequence of Helicobacter cinaedi Type Strain ATCC BAA-847. J. Bacteriol. 2012, 194, 5692. [Google Scholar] [CrossRef]
- The Human Microbiome Project Consortium. A Framework for Human Microbiome Research. Nature 2012, 486, 215–221.
- Charoenlap, N.; Shen, Z.; McBee, M.E.; Muthupalani, S.; Wogan, G.N.; Fox, J.G.; Schauer, D.B. Alkyl Hydroperoxide Reductase is Required for Helicobacter cinaedi Intestinal Colonization and Survival under Oxidative Stress in BALB/c and BALB/c Interleukin-10−/− mice. Infect. Immun. 2012, 80, 921–928. [Google Scholar] [CrossRef]
- Tseng, T.T.; Gratwick, K.S.; Kollman, J.; Park, D.; Nies, D.H.; Goffeau, A.; Saier, M.H., Jr. The RND Permease Superfamily: An Ancient, Ubiquitous and Diverse Family that Includes Human Disease and Development Proteins. J. Mol. Microbiol. Biotechnol. 1999, 1, 107–125. [Google Scholar]
- Morita, Y.; Kimura, N.; Mima, T.; Mizushima, T.; Tsuchiya, T. Roles of MexXY- and MexAB-Multidrug Efflux Pumps in Intrinsic Multidrug Resistance of Pseudomonas aeruginosa PAO1. J. Gen. Appl. Microbiol. 2001, 47, 27–32. [Google Scholar] [CrossRef]
- Poole, K.; Krebes, K.; McNally, C.; Neshat, S. Multiple Antibiotic Resistance in Pseudomonas aeruginosa: Evidence for Involvement of an Efflux Operon. J. Bacteriol. 1993, 175, 7363–7372. [Google Scholar]
- Lin, J.; Michel, L.O.; Zhang, Q. CmeABC Functions as a Multidrug Efflux System in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002, 46, 2124–2131. [Google Scholar]
- Trainor, E.A.; Horton, K.E.; Savage, P.B.; Testerman, T.L.; McGee, D.J. Role of the HefC Efflux Pump in Helicobacter pylori Cholesterol-dependent Resistance to Ceragenins and Bile Salts. Infect. Immun. 2011, 79, 88–97. [Google Scholar] [CrossRef]
- Srikumar, R.; Li, X.Z.; Poole, K. Inner Membrane Efflux Components are Responsible for Beta-Lactam Specificity of Multidrug Efflux Pumps in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 7875–7881. [Google Scholar]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Nishino, K.; Yamaguchi, A. Structures of the Multidrug Exporter AcrB Reveal a Proximal Multisite Drug-binding Pocket. Nature 2011, 480, 565–569. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Tsugawa, H.; Suzuki, H.; Muraoka, H.; Ikeda, F.; Hirata, K.; Matsuzaki, J.; Saito, Y.; Hibi, T. Enhanced bacterial efflux system is the first step to the development of metronidazole resistance in Helicobacter pylori. Biochem. Biophys. Res. Commun. 2011, 404, 656–660. [Google Scholar] [CrossRef]
- Francesco, V.D.; Zullo, A.; Hassan, C.; Giorgio, F.; Rosania, R.; Ierardi, E. Mechanisms of Helicobacter pylori Antibiotic Resistance: An Updated Appraisal. World J. Gastrointest. Pathophysiol. 2011, 2, 35–41. [Google Scholar] [CrossRef]
- Pumbwe, L.; Randall, L.P.; Woodward, M.J.; Piddock, L.J. Evidence for Multiple-antibiotic Resistance in Campylobacter jejuni not Mediated by CmeB or CmeF. Antimicrob. Agents Chemother. 2005, 49, 1289–1293. [Google Scholar] [CrossRef]
- Akiba, M.; Lin, J.; Barton, Y.W.; Zhang, Q. Interaction of CmeABC and CmeDEF in Conferring Antimicrobial Resistance and Maintaining Cell Viability in Campylobacter jejuni. J. Antimicrob. Chemother. 2006, 57, 52–60. [Google Scholar]
- Suerbaum, S.; Josenhans, C.; Sterzenbach, T.; Drescher, B.; Brandt, P.; Bell, M.; Droge, M.; Fartmann, B.; Fischer, H.P.; Ge, Z.; et al. The Complete Genome Sequence of the Carcinogenic Bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 2003, 100, 7901–7906. [Google Scholar]
- Cagliero, C.; Mouline, C.; Payot, S.; Cloeckaert, A. Involvement of the CmeABC Efflux Pump in the Macrolide Resistance of Campylobacter coli. J. Antimicrob. Chemother. 2005, 56, 948–950. [Google Scholar] [CrossRef]
- Yan, M.; Sahin, O.; Lin, J.; Zhang, Q. Role of the CmeABC Efflux Pump in the Emergence of Fluoroquinolone-resistant Campylobacter under Selection Pressure. J. Antimicrob. Chemother. 2006, 58, 1154–1159. [Google Scholar] [CrossRef]
- Lin, J.; Yan, M.; Sahin, O.; Pereira, S.; Chang, Y.J.; Zhang, Q. Effect of Macrolide Usage on Emergence of Erythromycin-resistant Campylobacter Isolates in Chickens. Antimicrob. Agents Chemother. 2007, 51, 1678–1686. [Google Scholar]
- Martin, F.A.; Posadas, D.M.; Carrica, M.C.; Cravero, S.L.; O'Callaghan, D.; Zorreguieta, A. Interplay Between Two RND Systems Mediating Antimicrobial Resistance in Brucella suis. J. Bacteriol. 2009, 191, 2530–2540. [Google Scholar] [CrossRef]
- Teran, W.; Felipe, A.; Segura, A.; Rojas, A.; Ramos, J.L.; Gallegos, M.T. Antibiotic-dependent Induction of Pseudomonas putida DOT-T1E TtgABC Efflux Pump is Mediated by the Drug Binding Repressor TtgR. Antimicrob. Agents Chemother. 2003, 47, 3067–3072. [Google Scholar]
- Hernould, M.; Gagne, S.; Fournier, M.; Quentin, C.; Arpin, C. Role of the AheABC Efflux Pump in Aeromonas hydrophila Intrinsic Multidrug Resistance. Antimicrob. Agents Chemother. 2008, 52, 1559–1563. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based Alignment Tool for Multiple Protein Sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef]
- McGee, D.J.; George, A.E.; Trainor, E.A.; Horton, K.E.; Hildebrandt, E.; Testerman, T.L. Cholesterol Enhances Helicobacter pylori Resistance to Antibiotics and ll-37. Antimicrob. Agents Chemother. 2011, 55, 2897–2904. [Google Scholar] [CrossRef]
- Lin, J.; Cagliero, C.; Guo, B.; Barton, Y.W.; Maurel, M.C.; Payot, S.; Zhang, Q. Bile Salts Modulate Expression of the CmeABC Multidrug Efflux Pump in Campylobacter jejuni. J. Bacteriol. 2005, 187, 7417–7424. [Google Scholar] [CrossRef]
- Lin, J.; Martinez, A. Effect of Efflux Pump Inhibitors on Bile Resistance and in Vivo Colonization of Campylobacter jejuni. J. Antimicrob. Chemother. 2006, 58, 966–972. [Google Scholar] [CrossRef]
- Lin, J.; Akiba, M.; Sahin, O.; Zhang, Q. CmeR Functions as a Transcriptional Repressor for the Multidrug Efflux Pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 2005, 49, 1067–1075. [Google Scholar] [CrossRef]
- Hwang, S.; Kim, M.; Ryu, S.; Jeon, B. Regulation of Oxidative Stress Response by CosR, an Essential Response Regulator in Campylobacter jejuni. PLoS One 2011, 6, e22300. [Google Scholar]
- Hwang, S.; Zhang, Q.; Ryu, S.; Jeon, B. Transcriptional Regulation of the CmeABC Multidrug Efflux Pump and the KatA Catalase by CosR in Campylobacter jejuni. J. Bacteriol. 2012, in press. [Google Scholar]
- Poole, K. Stress Responses as Determinants of Antimicrobial Resistance in Gram-negative Bacteria. Trends Microbiol. 2012, 20, 227–234. [Google Scholar] [CrossRef]
- Morita, Y.; Tomida, J.; Kawamura, Y. MexXY Multidrug Efflux System of Pseudomonas aeruginosa. Front. Microbiol. 2012, 3, e408. [Google Scholar]
- Hannenhalli, S.S.; Hayes, W.S.; Hatzigeorgiou, A.G.; Fickett, J.W. Bacterial Start Site Prediction. Nucleic Acids Res. 1999, 27, 3577–3582. [Google Scholar]
- Shine, J.; Dalgarno, L. The 3'-terminal Sequence of Escherichia coli 16S Ribosomal RNA: Complementarity to Nonsense Triplets and Ribosome Binding Sites. Proc. Natl. Acad. Sci. USA 1974, 71, 1342–1346. [Google Scholar] [CrossRef]
- Jeon, B.; Wang, Y.; Hao, H.; Barton, Y.W.; Zhang, Q. Contribution of CmeG to Antibiotic and Oxidative Stress Resistance in Campylobacter jejuni. J. Antimicrob. Chemother. 2011, 66, 79–85. [Google Scholar] [CrossRef]
- Van Amsterdam, K.; Bart, A.; van der Ende, A. A Helicobacter pylori TolC Efflux Pump Confers Resistance to Metronidazole. Antimicrob. Agents Chemother. 2005, 49, 1477–1482. [Google Scholar] [CrossRef]
- Chen, J.; Morita, Y.; Huda, M.N.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. VmrA, a Member of a Novel Class of Na(+)-coupled Multidrug Efflux Pumps from Vibrio parahaemolyticus. J. Bacteriol. 2002, 184, 572–576. [Google Scholar] [CrossRef]
- Miyamae, S.; Ueda, O.; Yoshimura, F.; Hwang, J.; Tanaka, Y.; Nikaido, H. A MATE Family Multidrug Efflux Transporter Pumps out Fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 2001, 45, 3341–3346. [Google Scholar] [CrossRef]
- Begum, A.; Rahman, M.M.; Ogawa, W.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. Gene Cloning and Characterization of Four MATE Family Multidrug Efflux Pumps from Vibrio cholerae Non-O1. Microbiol. Immunol. 2005, 49, 949–957. [Google Scholar]
- Masaoka, Y.; Ueno, Y.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. A Two-component Multidrug Efflux Pump, EbrAB, in Bacillus subtilis. J. Bacteriol. 2000, 182, 2307–2310. [Google Scholar] [CrossRef]
- Jack, D.L.; Storms, M.L.; Tchieu, J.H.; Paulsen, I.T.; Saier, M.H., Jr. A Broad-specificity Multidrug Efflux Pump Requiring a Pair of Homologous SMR-type Proteins. J. Bacteriol. 2000, 182, 2311–2313. [Google Scholar] [CrossRef]
- Nishino, K.; Nikaido, E.; Yamaguchi, A. Regulation and Physiological Function of Multidrug Efflux Pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 2009, 1794, 834–843. [Google Scholar] [CrossRef]
- Higashi, K.; Ishigure, H.; Demizu, R.; Uemura, T.; Nishino, K.; Yamaguchi, A.; Kashiwagi, K.; Igarashi, K. Identification of a Spermidine Excretion Protein Complex (MdtJI) in Escherichia coli. J. Bacteriol. 2008, 190, 872–878. [Google Scholar] [CrossRef]
- Imperi, F.; Tiburzi, F.; Visca, P. Molecular basis of Pyoverdine Siderophore Recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2009, 106, 20440–20445. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Morita, Y.; Tomida, J.; Kawamura, Y. Multidrug Efflux Systems in Helicobacter cinaedi. Antibiotics 2012, 1, 29-43. https://doi.org/10.3390/antibiotics1010029
Morita Y, Tomida J, Kawamura Y. Multidrug Efflux Systems in Helicobacter cinaedi. Antibiotics. 2012; 1(1):29-43. https://doi.org/10.3390/antibiotics1010029
Chicago/Turabian StyleMorita, Yuji, Junko Tomida, and Yoshiaki Kawamura. 2012. "Multidrug Efflux Systems in Helicobacter cinaedi" Antibiotics 1, no. 1: 29-43. https://doi.org/10.3390/antibiotics1010029




