Environmental and Public Health Implications of Water Reuse: Antibiotics, Antibiotic Resistant Bacteria, and Antibiotic Resistance Genes
Abstract
:1. Introduction
2. Antibiotics in Municipal Effluent
3. Antibiotic Resistant Bacteria and Associated Genes in Municipal Effluent
| Antibiotics | Gene class | Abundance in raw water | Abundance in final discharge/impacted environment | Treatment procedure | Geographical location | Ref. |
|---|---|---|---|---|---|---|
| Beta-lactam | ||||||
| blaTEM-uni | 106.15/mL sample 102.51/ng DNA 10−4.74/16S | 105.61/mL sample 102.38/ng DNA 10−3.64/16S | AS + Cl | Massachusetts, USA | [38] | |
| blaM-1 | 105.35/mL sample 101.10/ng DNA | 103.45/mL sample 102.03/ng DNA | AS + Cl | South Carolina, USA | [40] | |
| blavim | 10−1.22–101.26/ng DNA | ND–102.2/ng DNA | WWTP, not specified | Germany | [41] | |
| ampC | 100.34–102.66/ng DNA | 10−0.27–101.20/ng DNA | ||||
| ampC | NA | ND–101.84/mL sample | AS + P and N | Sabadell, Spain | [42] | |
| NA | ND/mL sample | Galatone WWTP | Nardò, Italy | |||
| NA | ND/mL sample | AS + UF + RO | Torreele, Belgium | |||
| blashv-5 | NA | ND–102.06/mL sample | AS + P and N | Sabadell, Spain | ||
| NA | ND/mL sample | Galatone WWTP | Nardò, Italy | |||
| NA | ND/mL sample | AS + UF + RO | Torreele, Belgium | |||
| mecA | NA | ND–102.75/mL sample | AS + P and N | Sabadell, Spain | ||
| NA | ND/mL sample | Galatone WWTP | Nardò, Italy | |||
| NA | ND/mL sample | AS + UF + RO | Torreele, Belgium | |||
| blaTEM | 104.4/mL sample | 103.8/mL sample | Not specified | Barcelona, Spain | [27] | |
| blaCTX-M | 103.3/mL sample | 102.1/mL sample | ||||
| mecA | 103.7/mL sample | 102.2/mL sample | ||||
| mecA | 101.7/mL sample 10−0.07/ng DNA | 100.7/mL sample 10−0.59/ng DNA | AS + TF | Gothenburg, Sweden | [43] | |
| Macrolide | ||||||
| ermB | NA | ND–104.42/mL sample | AS + P and N | Sabadell, Spain | [42] | |
| NA | ND–103.13/mL sample | Galatone WWTP | Nardò, Italy | |||
| NA | ND–103.28/mL sample | AS + UF + RO | Torreele, Belgium | |||
| ermB | ~109.70/mL sample ~10−2.30/16S | ~107.70/mL sample ~10−3.60/16S | Water solids, aerobic digestor for approximately 3 months | Minnesota, USA | [44] | |
| ermB | ~107.48/mL sample ~10−3.52/16S | ~102.3–104.48/mL sample ~ND–10−3.82/16S | Secondary effluent, activated sludge | Shafdan, Israel | [45] | |
| ermF | ~107.78/mL sample ~10−3.22/16S | ~103.48–105.3/mL sample ~10−3.05–10−2.52/16S | ||||
| Tetracycline | ||||||
| tetW | 105.37−107.4/mL sample ~10−3.12/16S | ND–103.63/mL sample ND/16S | AS/OD/RBCs/MBR + UV/Cl | Michigan, USA | [46] | |
| tetO | 105.51–107.61/mL sample ~10−3.12/16S | ND–103.96/mL sample ND/16S | ||||
| tetQ | 107.2–109/mL sample 105.3–106.8/ng DNA | 103.9–106.2/mL sample 103.7-105.4/ng DNA | AS/P and N/UV/Cl | Wisconsin, USA | [39] | |
| tetG | 106.4–107.8/mL sample 104.5–105.7/ng DNA | 104.2–105.9/mL sample 104–105/ng DNA | ||||
| tetC | 108.13–108.3/mL sample 105.45–105.65/ng DNA | ND–104.12/mL sample ND–103.57/ng DNA | AS + Cl | Hong Kong, China | [47] | |
| tetA | 107.78–108.2/mL sample 105.09–105.57/ng DNA | ND–104.33/mL sample ND–103.78/ng DNA | ||||
| tetA | 107.7/mL sample 105.55/ng DNA | 106.15/mL sample 105.24/ng DNA | AS | Nanjing, China | [26] | |
| tetC | 107.91/mL sample 105.76/ng DNA | 106.14/mL sample 105.23/ng DNA | ||||
| tetO | NA | ND–104.44/mL sample | AS + P and N | Sabadell, Spain | [42] | |
| NA | 102.93–104.58/mL sample | Galatone WWTP | Nardò, Italy | |||
| NA | ND–105.02/mL sample | AS + UF + RO | Torreele, Belgium | |||
| tetA | ~108.85/mL sample ~10−3.15/16S | ~107.85/mL sample ~10−3.46/16S | Water solids, aerobic digestor for approximately 3 months | Minnesota, USA | [44] | |
| tetW | ~109.78/mL sample ~10−2.22/16S | ~107.95/mL sample ~10−3.35/16S | ||||
| tetX | ~108.7/mL sample ~10−3.3/16S | ~109.48/mL sample ~10−1.82/16S | ||||
| tetO | NA | ~103, 101.7, 102/mL sample ~10−2.3, 10−2.6, 10−2.7/16S | Secondary effluent, chlorinated effluent, dechlorinated effluent | Western USA | [48] | |
| tetW | NA | ~103.6, 102.3,102/mL sample ~10−1.7, 10−2, 10−2.7/16S | ||||
| tetO | ~107.3/mL sample ~10−3.7/16S | ~ND−103/mL sample ~ND–10−4.4/16S | Secondary effluent, activated sludge | Shafdan, Israel | [45] | |
| tetM | 10−3.87–10−2.42/16S | NA | Rural domestic sewage treatment system, usually anaerobic digestor | Hangzhou, China | [49] | |
| tetO | 10−4.41–10−3.24/16S | |||||
| tetQ | 10−4.64–10−2.8/16S | |||||
| tetW | 10−3.16–10−2.03/16S | |||||
| tetM | ~10−2.70–10−2.30/16S | NA | Urban WWTP, usually oxidation ditch or anaerobic oxic zones | |||
| tetO | ~10−3.00–10−2.70/16S | |||||
| tetQ | ~10−2.82–10−2.00/16S | |||||
| tetW | ~10−1.70–10−1.30/16S | |||||
| Sulfonamide | ||||||
| sul-I | 105.46–107.54/mL sample ~10−3.4/16S | 104.37–106.75/mL sample ~10−3.4/16S | AS/OD/RBCs/MBR + UV/Cl | Michigan, USA | [46] | |
| sul-I | ~106.4/mL sample ~10−1.52/16S | ~106.5/mL sample ~10−1.1/16S | Not specified | Lausanne, Switzerland | [50] | |
| sul-II | ~105.6/mL sample ~10−2.3/16S | ~105.5/mL sample ~10−2/16S | ||||
| sul-I | ~108.90/mL sample ~10−3.10/16S | ~108/mL sample ~10−3.30/16S | Water solids, aerobic digestor for approximately 3 months | Minnesota, USA | [44] | |
| sul-I | NA | ~104.9, 103.7, 103.9/mL sample ~10−0.4, 10−0.6, 10−0.8/16S | Secondary effluent, chlorinated effluent, dechlorinated effluent | Western USA | [48] | |
| sul-II | NA | ~104.6, 102, 101.9/mL sample ~10−0.7, 10−2.3, 10−2.8/16S | ||||
| sul-I | ~108.48/mL sample ~10−2.4/16S | ~104.78–105.48/mL sample ~10−2.52–10−1.7/16S | Secondary effluent, activated sludge | Shafdan, Israel | [45] | |
| sul-II | ~108.30/mL sample ~10−2.7/16S | ~103.48–104.88/mL sample ~10−3.3–10−2.4/16S | ||||
| sul-I | ~10−2.70–10−1.70/16S | NA | Rural domestic sewage treatment system, usually anaerobic digestor | Hangzhou, China | [49] | |
| sul-II | ~10−2.52–10−1.15/16S | |||||
| sul-I | ~10−2.15–10−1.7/16S | NA | Urban WWTP, usually oxidation ditch or anaerobic oxic zones | |||
| sul-II | ~10−2.00–10−1.70/16S | |||||
| Others | ||||||
| vanA | <10−0.09/ng DNA | ND–10−2/ng DNA | WWTP, not specified | Germany | [41] | |
| vanA | NA | ND/mL sample | AS + P and N | Sabadell, Spain | [42] | |
| NA | ND/mL sample | Galatone WWTP | Nardò, Italy | |||
| NA | ND/mL sample | AS + UF + RO | Torreele, Belgium | |||
4. Antibiotics in Livestock Production Farms
5. Antibiotic Resistant Bacteria and Associated Genes in Livestock Production Farms
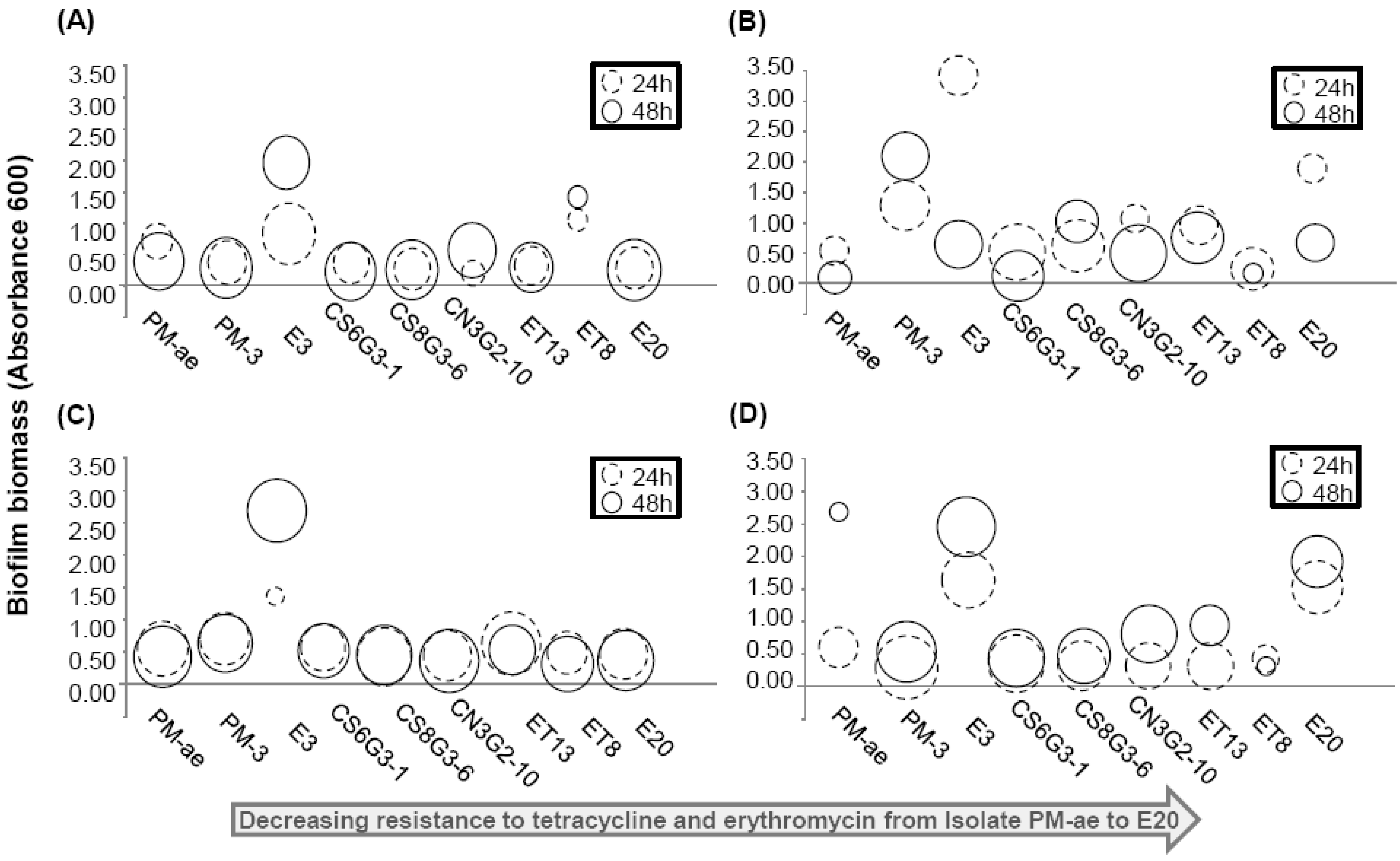
| Isolate name | Origin | Nearest match based on 16S rRNA gene (Max identity %) | MIC (µg/mL) | Motility * | ||||
|---|---|---|---|---|---|---|---|---|
| Tet | Erm | R2A | TYG | Nutrient | LB | |||
| PM-ae | Manure pit | Enterococcus avium (99%) JX185519 | 32 | 256 | N.A. | - | - | - |
| PM-3 | Manure pit | Uncultured bacterium clone (99%) JX185520 | 32 | 256 | - | - | - | - |
| E3 | Soil, 1 day after manure application | Staphylococcus simulans (99%) JX185521 | 32 | 256 | - | - | - | - |
| CS6G3-1 | Soil, 42 day after manure application | Achromobacter sp. (99%) JX185522 | 8 | 8 | + | + | + | + |
| CS8G3-6 | Soil, 42 day after manure application | Dyella sp. (99%) JX185523 | 8 | 4 | + | + | + | + |
| CN3G2-10 | Soil, 21 day after manure application | Uncultured Flexibacter sp. (99%) JX185524 | 16 | 16 | - | - | - | - |
| ET13 | Soil, 1 day after manure application | Burkholderia cenocepacia (99%) JX185525 | 4 | 16 | + | + | + | + |
| ET8 | Soil, 1 day after manure application | Brevibacillus brevis (99%) JX185526 | <4 | 4 | + | + | + | - |
| E20 | Soil, 1 day after manure application | Microbacterium takaoensis (99%) JX185527 | <4 | 4 | + | + | + | + |
| Antibiotics | Gene class | Abundance in the raw water | Abundance in the final discharge/impacted environment | Treatment procedure | Geographical location | Ref. |
|---|---|---|---|---|---|---|
| Macrolide | ||||||
| ermA | ND–10−3.15/16S | ND/16S | Lagoon Groundwater (Pig) | Illinois, USA | [80] | |
| ermB | 10−3.62–10−2/16S | ND–10−3.66/16S | ||||
| ermC | ND–10−3.06/16S | ND–10−3.68/16S | ||||
| Tetracycline | ||||||
| tetW | 10−3.2–10−3.0/16S | 10−6.2–10−4/16S | Lagoon Irrigation ditch (Dairy cattle) | Colorado, USA | [81] | |
| tetO | 10−3.9–10−3.5/16S | ND–10−5.2/16S | ||||
| tetM | 10−2.34–10−1.48/16S | ND–10−2.07/16S | Lagoon Groundwater (Pig) | Illinois, USA | [82] | |
| tetO | 10−2.74–10−1.34/16S | ND–10−3.04/16S | ||||
| tetQ | 10−1.93–10−0.92/16S | ND–10−2.07/16S | ||||
| tetW | 10−2.53–10−1.33/16S | ND–10−2.35/16S | ||||
| tetC | 10−2.85–10−1.15/16S | ND–10−0.99/16S | ||||
| tetH | <10−3.33–10−3.31/16S | ND–10−2.43/16S | ||||
| tetZ | <10−4.22–10−2.46/16S | ND–10−2.55/16S | ||||
| tetO | 104.1–105.2/mL sample | NA | Lagoon (Cattle) | Midwest, USA | [75] | |
| tetW | 105.1–105.5/mL sample | NA | ||||
| tetQ | 104.9–105.9/mL sample | NA | ||||
| tetA and tetC | 106.0–1010/g sample | 106.8–107.5/g sample | Effluent from confinement houses Lagoon | North Carolina or Ohio, USA | [83] | |
| tetG | 107.8–108.8/g sample | 107.3–108.4/g sample | ||||
| RPP (7 genes) | 107.8–109.2/g sample | 108.2–108.7/g sample | ||||
| tetM | NA | ~102.8– 106.1/mL sample ~10−5.7–10−2.2/16S | Cattle feedlot lagoons using different amount of antibiotics | Midwest, USA | [84] | |
| tetO | ~102.7–104.9/mL sample ~10−4.8–10−3/16S | |||||
| tetQ | ~103.2–105.5/mL sample ~10−5.7–10−3/16S | |||||
| tetW | ~102–104.5/mL sample ~10−4.8–10−3.1/16S | |||||
| tetB | ~102–103/mL sample ~10−5.6–10−5.3/16S | |||||
| tetL | ~100.4–101.8/mL sample ~10−6.7–10−5.2/16S | |||||
| tetW | NA | 10−2.4–10−1.8/16S | Water solids, Lagoon (Beef, dairy) | USA | [85] | |
| NA | ~10−1.9/16S | Water solids, Lagoon (swine) | ||||
| NA | ~10−4.8/16S | Water solids, Lagoon (chicken layer) | ||||
| tetQ | 10−2.43–10−2.21/16S 104.96–105.22/ng DNA | ND–10−1.93/16S ND–103.47/ng DNA | Lagoon (Pig) Groundwater | Illinois, USA | [77] | |
| tetZ | 10−4.39–10−3.84/16S 103.0–103.59/ng DNA | ND–10−2.28/16S ND–102.92/ng DNA | ||||
| Sulfonamide | ||||||
| sul–I | 10−1.5–10−1.4/16S | 10−2.6–10−1.6/16S | Lagoon Irrigation ditch (Animal) | Colorado, USA | [81] | |
| sul–II | 10−4.3–10−3.9/16S | ND/16S | ||||
| sul–I | NA | ~10−2.5–10−2/16S | Water solids, Lagoon (Beef, dairy) | USA | [85] | |
| sul–II | NA | ~10−1.4–10−1.1/16S | ||||
| sul–I | NA | ~10−1.4/16S | Water solids, Lagoon (swine) | |||
| sul–II | NA | ~10−0.2/16S | ||||
| sul–I | NA | ~10−2.7/16S | Water solids, Lagoon (chicken layer) | |||
| sul–II | NA | ~10−2.2/16S | ||||
| Others | ||||||
| cmlA | 104.8–106.05/mL sample 10−2.15–10−1.49/16S | NA | Lagoon (Swine feedlots) | Beijing, China | [74] | |
| floR | 104.94–106.05/mL sample 10−1.8–10−1.41/16S | |||||
| fexA | 104.52–106.21/mL sample 10−1.9–10−1.33/16S | |||||
| cfr | 104.86–106.1/mL sample 10−1.66–10−1.44/16S | |||||
| fexB | 104.9–106.2/mL sample 10−1.65–10−1.34/16S | |||||
| Type of ecosystem studied | Gene class | Type of antibiotics the gene class was resistant to | Abundance | Geographical location | Ref. |
|---|---|---|---|---|---|
| 7d soils subjected to one time batch irrigation, treated municipal effluent | sul-I | Sulfonamide | 101.85–103/g sample | Western USA | [48] |
| sul-II | 103–103.7/g sample | ||||
| tetO | Tetracycline | 101.6–101.7/g sample | |||
| tetW | 101.3–101.7/g sample | ||||
| 7d soils subjected to periodic irrigation, treated municipal effluent | sul-I | Sulfonamide | 102.3–102.95/g sample | ||
| sul-II | 101.48–103/g sample | ||||
| tetO | Tetracycline | 100.95–101.48/g sample | |||
| tetW | 101.48–103/g sample | ||||
| Irrigated soil, subjected to treated municipal wastewater irrigation | sul-I | Sulfonamide | ND–106.48/g sample ND–10−2.3/16S | Shafdan, Israel | [45] |
| sul-II | ND–105.3/g sample ND–10−3.52/16S | ||||
| ermB | Erythromycin | ND–103.95/g sample ND–10−5/16S | |||
| ermF | ND–105.85/g sample ND–10−3.05/16S | ||||
| tetO | Tetracycline | ND | |||
| Aquaculture, sediments | sul-I | Sulfonamide | 10−4.52–10−3.48/16S | Tianjin, China | [86] |
| sul-II | 10−3.7–10−2.74/16S | ||||
| tetW | Tetracycline | 10−4.96–10−3.36/16S | |||
| tetM | ND–10−3.7/16S | ||||
| tetO | ND–10−4/16S | ||||
| tetT | 10−6.67–10−5.51/16S | ||||
| Swine feedlot, soil | tetM | Tetracycline | 10−4.5–10−1.4/16S | Beijing, Tianjin, Jiaxing, China | [87] |
| tetO | 10−4.4–10−2.2/16S | ||||
| tetQ | 10−4.2–10−2/16S | ||||
| tetW | 10−4.8–10−2.2/16S | ||||
| tetT | ND–10−3.2/16S | ||||
| Swine, compost manure | sul-I | Sulfonamide | 10−1/16S | New Territories, Hong Kong | [88] |
| sul-II | 10−1.05/16S | ||||
| dfrA1 | 10−1.96/16S | ||||
| dfrA7 | 10−2.15/16S | ||||
| tetC | Tetracycline | 10−2.51/16S | |||
| tetG | 10−1.72/16S | ||||
| tetQ | 10−3.4/16S | ||||
| tetZ | 10−2.52/16S | ||||
| tetW | 10−3.62/16S | ||||
| tetY | 10−1.74/16S | ||||
| gyrA | Fluoroquinolone | 10−6/16S | |||
| parC | 10−6.35/16S | ||||
| Swine, manure-applied soil | tetQ | Tetracycline | ND–10−2.03/16S ND–104.53/ng DNA | Illinois, USA | [77] |
| tetZ | ND–10−3.39/16S ND–102.2/ng DNA | ||||
| Swine, manure-applied soil | cmlA | Chloramphenicol | 104.69–105.52/g sample 10−2.28–10−1.62/16S | Beijing, China | [74] |
| floR | 104.91–105.47/g sample 10−2.17–10−1.82/16S | ||||
| fexA | 104.88–105.42/g sample 10−2.34–10−1.65/16S | ||||
| cfr | 105.01–105.69/g sample 10−2.02–10−1.5/16S | ||||
| fexB | 105.06–105.61/g sample 10−2.03–10−1.61/16S |
6. Antibiotic Resistance Genes and Resistant Bacteria: Their Persistence and Fate
7. “Perfect Microbial Storm”
8. Conclusions
Acknowledgements
Conflict of Interest
References
- Jimenez, B. Water reuse: An International Survey of Current Practice, Issues and Needs; IWA Publishing: London, UK, 2008. [Google Scholar]
- Solley, W.B.; Pierce, R.R.; Perlman, H.A. Estimated Use of Water in the United States in 1990; Geological Survey: Washington, DC, USA, 1993. [Google Scholar]
- Asano, T.; Burton, F.L.; Leverenz, H.L.; Tsuchihashi, R.; Tchobanoglous, G. Water Reuse: Issues, Technologies, and Applications, 1st ed.; McGraw-Hill: New York, NY, USA, 2007; pp. 954–955. [Google Scholar]
- Chee-Sanford, J.C.; Mackie, R.I.; Koike, S.; Krapac, I.G.; Lin, Y.F.; Yannarell, A.C.; Maxwell, S.; Aminov, R.I. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 2009, 38, 1086–1108. [Google Scholar] [CrossRef]
- Vander Stichele, R.H.; Elseviers, M.M.; Ferech, M.; Blot, S.; Goossens, H. Hospital consumption of antibiotics in 15 European countries: Results of the ESAC retrospective data collection (1997–2002). J. Antimicrob. Chemother. 2006, 58, 159–167. [Google Scholar] [CrossRef]
- WHO. WHO Collaborating Centre for drug statistics methodology. Available online: http://www.whocc.no/atcddd/ (accessed on 30 May 2013).
- UCS, Hogging it: Estimates of Antimicrobial Abuse in Livestock; UCS Publishing: Cambridge, MA, USA, 2001; p. 109.
- Wise, R. Antimicrobial resistance: Priorities for action. J. Antimicrob. Chemother. 2002, 49, 585–586. [Google Scholar] [CrossRef]
- Ferreira da Silva, M.; Tiago, I.; Verissimo, A.; Boaventura, R.A.; Nunes, O.C.; Manaia, C.M. Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2006, 55, 322–329. [Google Scholar] [CrossRef]
- Observatorio Nacional de Saude. Medicos sentinela (ONSA). Available online: http://www.onsa.pt/ (accessed on 26 July 2013).
- Patrick, D.M.; Marra, F.; Hutchinson, J.; Monnet, D.L.; Ng, H.; Bowie, W.R. Per capita antibiotic consumption: How does a North American jurisdiction compare with Europe? Clin. Infect. Dis. 2004, 39, 11–17. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. 2010, 8, 423–435. [Google Scholar] [CrossRef]
- Asano, T.; Burton, F.L.; Leverenz, H.L.; Tsuchihashi, R.; Tchobanoglous, G. Water Reuse: Issues, Technologies, and Applications, 1st ed.; McGraw-Hill: New York, NY, USA, 2007; pp. 170–177. [Google Scholar]
- Smolinski, M.S.; Hamburg, M.A.; Lederberg, J. Microbial Threats to Health: Emergence, Detection, and Response; National Academies Press: Washington, DC, USA, 2003; p. 367. [Google Scholar]
- Michael, I.; Rizzo, L.; McArdell, C.S.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–995. [Google Scholar] [CrossRef]
- Senta, I.; Terzic, S.; Ahel, M. Occurrence and fate of dissolved and particulate antimicrobials in municipal wastewater treatment. Water Res. 2013, 47, 705–714. [Google Scholar] [CrossRef]
- Rogers, H.R. Sources, behaviour and fate of organic contaminants during sewage treatment and in sewage sludges. Sci. Total Environ. 1996, 185, 3–26. [Google Scholar] [CrossRef]
- Clara, M.; Strenn, B.; Gans, O.; Martinez, E.; Kreuzinger, N.; Kroiss, H. Removal of selected pharmaceuticals, fragrances and endocrine disrupting compounds in a membrane bioreactor and conventional wastewater treatment plants. Water Res. 2005, 39, 4797–4807. [Google Scholar] [CrossRef]
- Watkinson, A.J.; Murby, E.J.; Costanzo, S.D. Removal of antibiotics in conventional and advanced wastewater treatment: Implications for environmental discharge and wastewater recycling. Water Res. 2007, 41, 4164–4176. [Google Scholar] [CrossRef]
- Gobel, A.; McArdell, C.S.; Joss, A.; Siegrist, H.; Giger, W. Fate of sulfonamides, macrolides, and trimethoprim in different wastewater treatment technologies. Sci. Total Environ. 2007, 372, 361–371. [Google Scholar] [CrossRef]
- Eddy, M.; Tchobanoglous, G.; Burton, F.L.; Stensel, H.D. Wastewater Engineering: Treatment and Reuse, 4th ed.; McGraw-Hill Higher Education: New York, NY, USA, 2002; p. 1408. [Google Scholar]
- Merlin, C.; Bonot, S.; Courtois, S.; Block, J.C. Persistence and dissemination of the multiple-antibiotic-resistance plasmid pB10 in the microbial communities of wastewater sludge microcosms. Water Res. 2011, 45, 2897–2905. [Google Scholar] [CrossRef]
- Rysz, M.; Mansfield, W.R.; Fortner, J.D.; Alvarez, P.J. Tetracycline resistance gene maintenance under varying bacterial growth rate, substrate and oxygen availability, and tetracycline concentration. Environ. Sci. Technol. 2013, 47, 6995–7001. [Google Scholar]
- Salyers, A.A.; Gupta, A.; Wang, Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004, 12, 412–416. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, B.; Zhang, Y.; Zhang, T.; Yang, L.; Fang, H.H.; Ford, T.; Cheng, S. Class 1 integronase gene and tetracycline resistance genes tetA and tetC in different water environments of Jiangsu province, China. Ecotoxicology 2009, 18, 652–660. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One 2011, 6, e17549. [Google Scholar] [CrossRef]
- Zhang, X.X.; Zhang, T. Occurrence, abundance, and diversity of tetracycline resistance genes in 15 sewage treatment plants across China and other global locations. Environ. Sci. Technol. 2011, 45, 2598–2604. [Google Scholar] [CrossRef]
- Zhang, Y.; Marrs, C.F.; Simon, C.; Xi, C. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 2009, 407, 3702–3706. [Google Scholar] [CrossRef]
- Guardabassi, L.; Lo Fo Wong, D.M.; Dalsgaard, A. The effects of tertiary wastewater treatment on the prevalence of antimicrobial resistant bacteria. Water Res. 2002, 36, 1955–1964. [Google Scholar] [CrossRef]
- Ferreira da Silva, M.; Vaz-Moreira, I.; Gonzalez-Pajuelo, M.; Nunes, O.C.; Manaia, C.M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2007, 60, 166–176. [Google Scholar] [CrossRef]
- Huang, J.J.; Hu, H.Y.; Tang, F.; Li, Y.; Lu, S.Q.; Lu, Y. Inactivation and reactivation of antibiotic-resistant bacteria by chlorination in secondary effluents of a municipal wastewater treatment plant. Water Res. 2011, 45, 2775–2781. [Google Scholar] [CrossRef]
- Tamaki, H.; Sekiguchi, Y.; Hanada, S.; Nakamura, K.; Nomura, N.; Matsumura, M.; Kamagata, Y. Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl. Environ. Microbiol. 2005, 71, 2162–2169. [Google Scholar] [CrossRef]
- Staley, J.T.; Konopka, A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Ann. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar]
- Bush, K. New beta-lactamases in gram-negative bacteria: Diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 2001, 32, 1085–1089. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Medeiros, A.A. More extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 1991, 35, 1697–1704. [Google Scholar] [CrossRef]
- Lachmayr, K.L.; Kerkhof, L.J.; Dirienzo, A.G.; Cavanaugh, C.M.; Ford, T.E. Quantifying nonspecific TEM beta-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 2009, 75, 203–211. [Google Scholar] [CrossRef]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef]
- Uyaguari, M.I.; Fichot, E.B.; Scott, G.I.; Norman, R.S. Characterization and quantitation of a novel beta-lactamase gene found in a wastewater treatment facility and the surrounding coastal ecosystem. Appl. Environ. Microbiol. 2011, 77, 8226–8233. [Google Scholar] [CrossRef]
- Schwartz, T. Strategies to assess and minimize the biological risk of antibiotic resistance in the environment. In Antimicrobial Resistance in the Environment; Keen, P., Montforts, M., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2012; pp. 251–264. [Google Scholar]
- Bockelmann, U.; Dorries, H.H.; Ayuso-Gabella, M.N.; Salgot de Marcay, M.; Tandoi, V.; Levantesi, C.; Masciopinto, C.; van Houtte, E.; Szewzyk, U.; Wintgens, T.; et al. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three european artificial groundwater recharge systems. Appl. Environ. Microbiol. 2009, 75, 154–163. [Google Scholar] [CrossRef]
- Borjesson, S.; Melin, S.; Matussek, A.; Lindgren, P.E. A seasonal study of the mecA gene and Staphylococcus aureus including methicillin-resistant S. aureus in a municipal wastewater treatment plant. Water Res. 2009, 43, 925–932. [Google Scholar] [CrossRef]
- Burch, T.R.; Sadowsky, M.J.; Lapara, T.M. Aerobic digestion reduces the quantity of antibiotic resistance genes in residual municipal wastewater solids. Front. Microbiol. 2013, 4, e17. [Google Scholar]
- Negreanu, Y.; Pasternak, Z.; Jurkevitch, E.; Cytryn, E. Impact of treated wastewater irrigation on antibiotic resistance in agricultural soils. Environ. Sci. Technol. 2012, 46, 4800–4808. [Google Scholar] [CrossRef]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, M.; Zhang, X.; Fang, H.H. Tetracycline resistance genes and tetracycline resistant lactose-fermenting Enterobacteriaceae in activated sludge of sewage treatment plants. Environ. Sci. Technol. 2009, 43, 3455–3460. [Google Scholar] [CrossRef]
- Fahrenfeld, N.; Ma, Y.; O'Brien, M.; Pruden, A. Reclaimed water as a reservoir of antibiotic resistance genes: Distribution system and irrigation implications. Front. Microbiol. 2013, 4, e130. [Google Scholar]
- Chen, H.; Zhang, M. Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China. Environ. Int. 2013, 55, 9–14. [Google Scholar] [CrossRef]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Burgmann, H. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbiol. 2012, 3, e106. [Google Scholar]
- KICP, The KAUST Industry Collaboration Program (KICP) Annual Strategic Study—Promoting Wastewater Reclamation and Reuse in the Kingdom of Saudi Arabia: Technology Trends, Innovation Needs, and Business Opportunities; KAUST: Thuwal, Saudi Arabia, 2010.
- USFDA Summary report on antimicrobials sold or distributed for use in food-producing animals. Available online: http://www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/UCM338170.pdf (accessed on 26 July 2013).
- Compassion in World Farming. Antibiotics in farm animal production: Public health and animal welfare. Available online: http://www.ciwf.org.uk/includes/documents/cm_docs/2011/a/antibiotics_in_animal_farming.pdf (accessed on 21 April 2013).
- DANMAP. Danmap 2011-use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. 2011. Available online: http://danmap.org/Downloads/~/media/Projekt%20sites/Danmap/DANMAP%20reports/Danmap_2011.ashx/ (accessed on 26 July 2013).
- Cromwell, G.L. Why and how antibiotics are used in swine production. Anim. Biotechnol. 2002, 13, 7–27. [Google Scholar] [CrossRef]
- Pei, R.; Cha, J.; Carlson, K.H.; Pruden, A. Response of antibiotic resistance genes (ARG) to biological treatment in dairy lagoon water. Environ. Sci. Technol. 2007, 41, 5108–5113. [Google Scholar] [CrossRef]
- Hong, P.Y.; Li, X.; Yang, X.; Shinkai, T.; Zhang, Y.; Wang, X.; Mackie, R.I. Monitoring airborne biotic contaminants in the indoor environment of pig and poultry confinement buildings. Environ. Microbiol. 2012, 14, 1420–1431. [Google Scholar] [CrossRef]
- Aga, D.S.; Goldfish, R.; Kulshrestha, P. Application of ELISA in determining the fate of tetracyclines in land-applied livestock wastes. Analyst 2003, 128, 658–662. [Google Scholar] [CrossRef]
- Aga, D.S.; O'Connor, S.; Ensley, S.; Payero, J.O.; Snow, D.; Tarkalson, D. Determination of the persistence of tetracycline antibiotics and their degradates in manure-amended soil using enzyme-linked immunosorbent assay and liquid chromatography-mass spectrometry. J. Agric. Food Chem. 2005, 53, 7165–7171. [Google Scholar]
- Cho, I.; Yamanishi, S.; Cox, L.; Methe, B.A.; Zavadil, J.; Li, K.; Gao, Z.; Mahana, D.; Raju, K.; Teitler, I.; et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2012, 488, 621–626. [Google Scholar] [CrossRef]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Flint, H.J. Microbiology: Antibiotics and adiposity. Nature 2012, 488, 601–602. [Google Scholar] [CrossRef]
- Hong, P.Y.; Croix, J.A.; Greenberg, E.; Gaskins, H.R.; Mackie, R.I. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS One 2011, 6, e25042. [Google Scholar]
- Hong, P.Y.; Wheeler, E.; Cann, I.K.; Mackie, R.I. Phylogenetic analysis of the fecal microbial community in herbivorous land and marine iguanas of the Galápagos islands using 16S rRNA-based pyrosequencing. ISME J. 2011, 5, 1461–1470. [Google Scholar] [CrossRef]
- Cotta, M.A.; Whitehead, T.R.; Zeltwanger, R.L. Isolation, characterization and comparison of bacteria from swine faeces and manure storage pits. Environ. Microbiol. 2003, 5, 737–745. [Google Scholar] [CrossRef]
- Jindal, A.; Kocherginskaya, S.; Mehboob, A.; Robert, M.; Mackie, R.I.; Raskin, L.; Zilles, J.L. Antimicrobial use and resistance in swine waste treatment systems. Appl. Environ. Microbiol. 2006, 72, 7813–7820. [Google Scholar] [CrossRef]
- Jiang, H.X.; Lu, D.H.; Chen, Z.L.; Wang, X.M.; Chen, J.R.; Liu, Y.H.; Liao, X.P.; Liu, J.H.; Zeng, Z.L. High prevalence and widespread distribution of multi-resistant Escherichia coli isolates in pigs and poultry in China. Vet. J. 2011, 187, 99–103. [Google Scholar] [CrossRef]
- Burgos, J.M.; Ellington, B.A.; Varela, M.F. Presence of multidrug-resistant enteric bacteria in dairy farm topsoil. J. Dairy Sci. 2005, 88, 1391–1398. [Google Scholar] [CrossRef]
- Aminov, R.I.; Chee-Sanford, J.C.; Garrigues, N.; Teferedegne, B.; Krapac, I.J.; White, B.A.; Mackie, R.I. Development, validation, and application of pcr primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 2002, 68, 1786–1793. [Google Scholar] [CrossRef]
- Aminov, R.I.; Garrigues-Jeanjean, N.; Mackie, R.I. Molecular ecology of tetracycline resistance: Development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 2001, 67, 22–32. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Ghosh, A.; Dowd, S.E.; Zurek, L. Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS One 2011, 6, e22451. [Google Scholar] [CrossRef]
- Li, J.; Shao, B.; Shen, J.; Wang, S.; Wu, Y. Occurrence of chloramphenicol-resistance genes as environmental pollutants from swine feedlots. Environ. Sci. Technol. 2013, 47, 2892–2897. [Google Scholar] [CrossRef]
- Smith, M.S.; Yang, R.K.; Knapp, C.W.; Niu, Y.; Peak, N.; Hanfelt, M.M.; Galland, J.C.; Graham, D.W. Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl. Environ. Microbiol. 2004, 70, 7372–7377. [Google Scholar] [CrossRef]
- O'Brien, T.F. Emergence, spread, and environmental effect of antimicrobial resistance: How use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 2002, 34, S78–S84. [Google Scholar] [CrossRef]
- Hong, P.Y.; Yannarell, A.C.; Dai, Q.; Ekizoglu, M.; Mackie, R.I. Monitoring the perturbation of soil and groundwater microbial communities due to pig production activities. Appl. Environ. Microbiol. 2013, 79, 2620–2629. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Aminov, R.I.; Krapac, I.J.; Garrigues-Jeanjean, N.; Mackie, R.I. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 2001, 67, 1494–1502. [Google Scholar] [CrossRef]
- Hong, P.Y.; Wu, J.H.; Liu, W.T. A high-throughput and quantitative hierarchical oligonucleotide primer extension (HOPE)-based approach to identify sources of faecal contamination in water bodies. Environ. Microbiol. 2009, 11, 1672–1681. [Google Scholar] [CrossRef]
- Koike, S.; Aminov, R.I.; Yannarell, A.C.; Gans, H.D.; Krapac, I.G.; Chee-Sanford, J.C.; Mackie, R.I. Molecular ecology of macrolide-lincosamide-streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Microbial. Ecol. 2010, 59, 487–498. [Google Scholar] [CrossRef]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef]
- Koike, S.; Krapac, I.G.; Oliver, H.D.; Yannarell, A.C.; Chee-Sanford, J.C.; Aminov, R.I.; Mackie, R.I. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period. Appl. Environ. Microbiol. 2007, 73, 4813–4823. [Google Scholar] [CrossRef]
- Yu, Z.; Michel, F.C., Jr.; Hansen, G.; Wittum, T.; Morrison, M. Development and application of real-time pcr assays for quantification of genes encoding tetracycline resistance. Appl. Environ. Microbiol. 2005, 71, 6926–6933. [Google Scholar] [CrossRef]
- Peak, N.; Knapp, C.W.; Yang, R.K.; Hanfelt, M.M.; Smith, M.S.; Aga, D.S.; Graham, D.W. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 2007, 9, 143–151. [Google Scholar] [CrossRef]
- McKinney, C.W.; Loftin, K.A.; Meyer, M.T.; Davis, J.G.; Pruden, A. Tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ. Sci. Technol. 2010, 44, 6102–6109. [Google Scholar] [CrossRef]
- Gao, P.; Mao, D.; Luo, Y.; Wang, L.; Xu, B.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012, 46, 2355–2364. [Google Scholar] [CrossRef]
- Wu, N.; Qiao, M.; Zhang, B.; Cheng, W.D.; Zhu, Y.G. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ. Sci. Technol. 2010, 44, 6933–6939. [Google Scholar] [CrossRef]
- Selvam, A.; Xu, D.; Zhao, Z.; Wong, J.W. Fate of tetracycline, sulfonamide and fluoroquinolone resistance genes and the changes in bacterial diversity during composting of swine manure. Bioresour. Technol. 2012, 126, 383–390. [Google Scholar] [CrossRef]
- Akiyama, T.; Savin, M.C. Populations of antibiotic-resistant coliform bacteria change rapidly in a wastewater effluent dominated stream. Sci. Total Environ. 2010, 408, 6192–6201. [Google Scholar] [CrossRef]
- Crecchio, C.; Stotzky, G. Binding of DNA on humic acids: Effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol. Biochem. 1998, 30, 1061–1067. [Google Scholar] [CrossRef]
- Khanna, M.; Stotzky, G. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl. Environ. Microbiol. 1992, 58, 1930–1939. [Google Scholar]
- Romanowski, G.; Lorenz, M.G.; Wackernagel, W. Plasmid DNA in a groundwater aquifer microcosm-adsorption, DNase resistance and natural genetic transformation of Bacillus subtilis. Mol. Ecol. 1993, 2, 171–181. [Google Scholar] [CrossRef]
- Lorenz, M.G.; Wackernagel, W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 1994, 58, 563–602. [Google Scholar]
- Lu, N.; Zilles, J.L.; Nguyen, T.H. Adsorption of extracellular chromosomal DNA and its effects on natural transformation of Azotobacter vinelandii. Appl. Environ. Microbiol. 2010, 76, 4179–4184. [Google Scholar] [CrossRef]
- McKinney, C.W.; Pruden, A. Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ. Sci. Technol. 2012, 46, 13393–13400. [Google Scholar] [CrossRef]
- USEPA. Ultraviolet disinfection guidance manual for the final long term 2 enhanced surface water treatment rule. 2006. Available online: http://www.epa.gov/ogwdw/disinfection/lt2/pdfs/guide_lt2_uvguidance.pdf (accessed on 26 July 2013).
- Bae, S.; Wuertz, S. Survival of host-associated bacteroidales cells and their relationship with Enterococcus spp., Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and adenovirus in freshwater microcosms as measured by propidium monoazide-quantitative PCR. Appl. Environ. Microbiol. 2012, 78, 922–932. [Google Scholar] [CrossRef]
- Jeanneau, L.; Solecki, O.; Wery, N.; Jarde, E.; Gourmelon, M.; Communal, P.Y.; Jadas-Hecart, A.; Caprais, M.P.; Gruau, G.; Pourcher, A.M. Relative decay of fecal indicator bacteria and human-associated markers: A microcosm study simulating wastewater input into seawater and freshwater. Environ. Sci. Technol. 2012, 46, 2375–2382. [Google Scholar] [CrossRef] [Green Version]
- Walters, S.P.; Yamahara, K.M.; Boehm, A.B. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: Implications for their use in assessing risk in recreational waters. Water Res. 2009, 43, 4929–4939. [Google Scholar] [CrossRef]
- Szczepanowski, R.; Eikmeyer, F.; Harfmann, J.; Blom, J.; Rogers, L.M.; Top, E.M.; Schluter, A. Sequencing and comparative analysis of IncP-1alpha antibiotic resistance plasmids reveal a highly conserved backbone and differences within accessory regions. J. Biol. Technol. 2011, 155, 95–103. [Google Scholar]
- Haines, A.S.; Jones, K.; Batt, S.M.; Kosheleva, I.A.; Thomas, C.M. Sequence of plasmid pBS228 and reconstruction of the IncP-1alpha phylogeny. Plasmid 2007, 58, 76–83. [Google Scholar] [CrossRef]
- Pansegrau, W.; Lanka, E.; Barth, P.T.; Figurski, D.H.; Guiney, D.G.; Haas, D.; Helinski, D.R.; Schwab, H.; Stanisich, V.A.; Thomas, C.M. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 1994, 239, 623–663. [Google Scholar] [CrossRef]
- Tennstedt, T.; Szczepanowski, R.; Krahn, I.; Puhler, A.; Schluter, A. Sequence of the 68,869 bp IncP-1alpha plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 2005, 53, 218–238. [Google Scholar] [CrossRef]
- Eikmeyer, F.; Hadiati, A.; Szczepanowski, R.; Wibberg, D.; Schneiker-Bekel, S.; Rogers, L.M.; Brown, C.J.; Top, E.M.; Puhler, A.; Schluter, A. The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid 2012, 68, 13–24. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.A.; Alm, E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef]
- Sommer, M.O.; Dantas, G.; Church, G.M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 2009, 325, 1128–1131. [Google Scholar] [CrossRef]
- Cogliani, C.; Goossens, H.; Greko, C. Restricting antimicrobial use in food animals: Lessons from Europe. Microbes 2011, 6, 274–279. [Google Scholar]
- Bager, F.; Aarestrup, F.M.; Madsen, M.; Wegener, H.C. Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microb. Drug Resistance 1999, 5, 53–56. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Seyfarth, A.M.; Emborg, H.D.; Pedersen, K.; Hendriksen, R.S.; Bager, F. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 2001, 45, 2054–2059. [Google Scholar] [CrossRef]
- MARAN, Monitoring of antimicrobial resistance and antibiotic usage in animals in the Netherlands. 2012. Available online: http://www.uu.nl/SiteCollectionImages/Fac_DGK/Nieuwsplaatjes/Nieuws/2012/NethmapMaran_Web.pdf (accessed on 26 July 2013).
- Borjesson, S.; Egervarn, M.; Lindblad, M.; Englund, S. Frequent occurrence of extended-spectrum beta-lactamase- and transferable ampC beta-lactamase-producing Escherichia coli on domestic chicken meat in Sweden. Appl. Environ. Microbiol. 2013, 79, 2463–2466. [Google Scholar] [CrossRef]
- Travers, K.; Barza, M. Morbidity of infections caused by antimicrobial-resistant bacteria. Clin. Infect. Dis. 2002, 34, S131–S134. [Google Scholar] [CrossRef]
- Kumar, K.; Gupta, S.C.; Baidoo, S.K.; Chander, Y.; Rosen, C.J. Antibiotic uptake by plants from soil fertilized with animal manure. J. Environ. Qual. 2005, 34, 2082–2085. [Google Scholar] [CrossRef]
- Dolliver, H.; Kumar, K.; Gupta, S. Sulfamethazine uptake by plants from manure-amended soil. J. Environ. Qual. 2007, 36, 1224–1230. [Google Scholar] [CrossRef]
- Boxall, A.B.; Johnson, P.; Smith, E.J.; Sinclair, C.J.; Stutt, E.; Levy, L.S. Uptake of veterinary medicines from soils into plants. J. Agric. Food Chem. 2006, 54, 2288–2297. [Google Scholar] [CrossRef]
- Wu, C.; Spongberg, A.L.; Witter, J.D.; Fang, M.; Czajkowski, K.P. Uptake of pharmaceutical and personal care products by soybean plants from soils applied with biosolids and irrigated with contaminated water. Environ. Sci. Technol. 2010, 44, 6157–6161. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hong, P.-Y.; Al-Jassim, N.; Ansari, M.I.; Mackie, R.I. Environmental and Public Health Implications of Water Reuse: Antibiotics, Antibiotic Resistant Bacteria, and Antibiotic Resistance Genes. Antibiotics 2013, 2, 367-399. https://doi.org/10.3390/antibiotics2030367
Hong P-Y, Al-Jassim N, Ansari MI, Mackie RI. Environmental and Public Health Implications of Water Reuse: Antibiotics, Antibiotic Resistant Bacteria, and Antibiotic Resistance Genes. Antibiotics. 2013; 2(3):367-399. https://doi.org/10.3390/antibiotics2030367
Chicago/Turabian StyleHong, Pei-Ying, Nada Al-Jassim, Mohd Ikram Ansari, and Roderick I. Mackie. 2013. "Environmental and Public Health Implications of Water Reuse: Antibiotics, Antibiotic Resistant Bacteria, and Antibiotic Resistance Genes" Antibiotics 2, no. 3: 367-399. https://doi.org/10.3390/antibiotics2030367






