Dynamic Post-Transcriptional Regulation of HIV-1 Gene Expression
Abstract
:1. Introduction
2. Spatial and Temporal Definition of HIV-1 Transcription
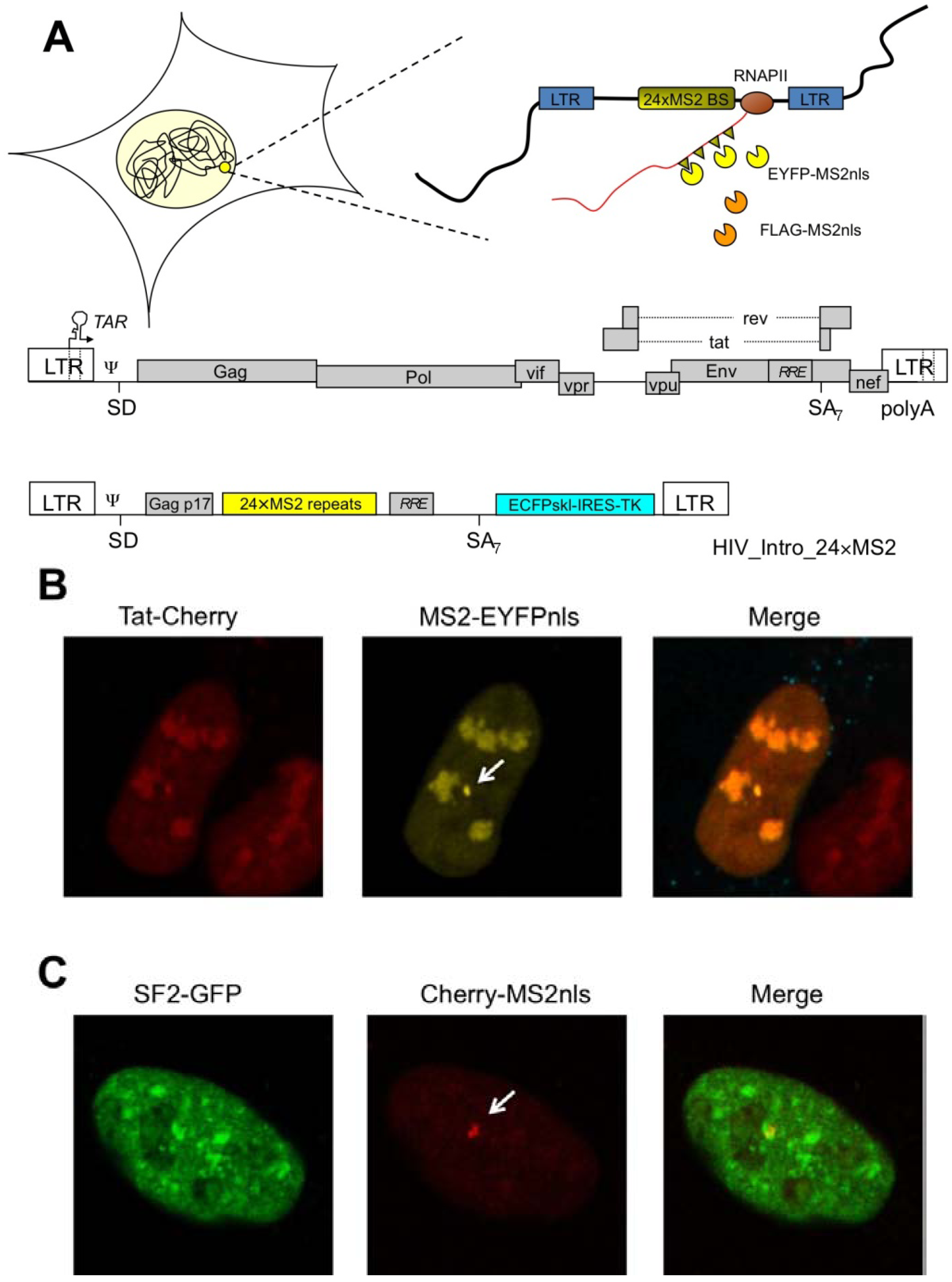
3. A Portfolio of HIV-1 RNA Species
4. HIV-1 Rev & Friends
| Cellular Protein | Proposed function in Rev Activity | References |
|---|---|---|
| Following the Order of Their Mention in the Text. | ||
| CK2 | Interacts with and phosphorylates Rev | [59] |
| PRMT6 | Methylates Rev | [60] |
| Crm-1 | Nuclear export or Rev-bound RRE containing RNAs | [64] |
| Importin-β | Import of Rev into the nucleus | [65,66] |
| DDX-3 | Helicase that promotes nuclear export of Rev-containing RNAs | [67] |
| DDX-1 | Helicase similar to DDX-3 but restricted to astrocytes | [68] |
| DDX-9 (RHA) | Helicase involved in remodeling RNA upstream of Rev | [69] |
| DDX-24 | Helicase that binds Rev and is involved in viral RNA packaging | [70] |
| B23 | Binds Rev and colocalizes to nulceoli | [71] |
| p32 | Human homologue of yeast YL2 | [72] |
| NAP-1 | Interacts with Rev nls | [73] |
| HIC | Binds Rev in the cytoplasm and modulates nuclear import through nls binding | [74] |
| eIF-5A | Binds Rev and is involved in nuclear export of viral RNA | [75,76] |
| PABP1 | Associates with HIV-1 RNAs in a Rev-dependent manner | [77] |
| hRIP | Binds Rev and promotes release of RNA from the nucleus | [78,79] |
| Sam68 | Complements Rev activity | [80,81] |
| Hax-1 | Inhibits Rev function in the cytoplasm | [82] |
| Following the Order of Their Mention in the Text. | ||
| PIMT | 5’-CAP modification of Rev-dependent transcripts | [62] |
| MATR3 | Promotes the nuclear export of Rev-dependent RNAs | [35,83] |
| SF2/ASF | Binds RRE in a Rev-dependent manner | [84] |
| Not Described in the Text. | ||
| RREBP49 | hnRNP F homologue that binds RRE | [85] |
| Pur α | Interacts with Rev and RRE | [86] |
| Prothymosin α | Interacts with Rev | [87] |
| NF90 | Inhibits Rev function | [88] |
| IkB | Negatively regulates Rev function | [89] |
| hnRNPA1 | Binds to repressor sequences in gag and stimulates Rev function | [53] |
| 16.4.1 | Interacts with Rev and Crm-1 | [90] |
| ATM | Enhances Rev function and viral replication | [91] |
| β-actin | Involved in Rev activity on nuclear export, together with eIF-5A, or translation | [92,93] |
5. From Transcription to Nuclear Export: The Journey of HIV-1 RRE-Containing RNAs
6. Conclusions
Acknowledgments
References
- Greene, W.C.; Peterlin, B.M. Charting HIV’s remarkable voyage through the cell: Basic science as a passport to future therapy. Nat. Med. 2002, 8, 673–680. [Google Scholar] [CrossRef]
- Marcello, A.; Lusic, M.; Pegoraro, G.; Pellegrini, V.; Beltram, F.; Giacca, M. Nuclear organization and the control of HIV-1 transcription. Gene 2004, 326, 1–11. [Google Scholar] [CrossRef]
- Nekhai, S.; Jeang, K.T. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: Role of cellular factors for Tat and Rev. Future Microbiol. 2006, 1, 417–426. [Google Scholar] [CrossRef]
- Karn, J.; Stoltzfus, C.M. Transcriptional and Posttranscriptional Regulation of HIV-1 Gene Expression. Cold Spring Harb. Perspect. Med. 2012, 2, 1–17. [Google Scholar]
- Karn, J. The molecular biology of HIV latency: Breaking and restoring the Tat-dependent transcriptional circuit. Curr. Opin. HIV AIDS 2011, 6, 4–11. [Google Scholar] [CrossRef]
- Han, Y.; Wind-Rotolo, M.; Yang, H.C.; Siliciano, J.D.; Siliciano, R.F. Experimental approaches to the study of HIV-1 latency. Nat. Rev. Microbiol. 2007, 5, 95–106. [Google Scholar] [CrossRef]
- Marcello, A. Latency: The hidden HIV-1 challenge. Retrovirology 2006, 3, 7. [Google Scholar] [CrossRef]
- Dinoso, J.B.; Kim, S.Y.; Wiegand, A.M.; Palmer, S.E.; Gange, S.J.; Cranmer, L.; O’Shea, A.; Callender, M.; Spivak, A.; Brennan, T.; et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2009, 106, 9403–9408. [Google Scholar]
- Yukl, S.A.; Shergill, A.K.; McQuaid, K.; Gianella, S.; Lampiris, H.; Hare, C.B.; Pandori, M.; Sinclair, E.; Gunthard, H.F.; Fischer, M.; et al. Effect of raltegravir-containing intensification on HIV burden and T-cell activation in multiple gut sites of HIV-positive adults on suppressive antiretroviral therapy. AIDS 2010, 24, 2451–2460. [Google Scholar]
- Gandhi, R.T.; Zheng, L.; Bosch, R.J.; Chan, E.S.; Margolis, D.M.; Read, S.; Kallungal, B.; Palmer, S.; Medvik, K.; Lederman, M.M.; et al. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: A randomized controlled trial. PLoS Med. 2010, 7, 8. [Google Scholar]
- Palmer, S.; Josefsson, L.; Coffin, J.M. HIV reservoirs and the possibility of a cure for HIV infection. J. Intern. Med. 2011, 270, 550–560. [Google Scholar] [CrossRef]
- Colin, L.; van Lint, C. Molecular control of HIV-1 postintegration latency: Implications for the development of new therapeutic strategies. Retrovirology 2009, 6, 111. [Google Scholar] [CrossRef]
- Stoltzfus, C.M. Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv. Virus Res. 2009, 74, 1–40. [Google Scholar] [CrossRef]
- Cochrane, A.W.; McNally, M.T.; Mouland, A.J. The retrovirus RNA trafficking granule: From birth to maturity. Retrovirology 2006, 3, 18. [Google Scholar] [CrossRef]
- Ott, M.; Geyer, M.; Zhou, Q. The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host Microbe 2012, 10, 426–435. [Google Scholar]
- Peterlin, B.M.; Price, D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 2006, 23, 297–305. [Google Scholar] [CrossRef]
- Jeang, K.T.; Xiao, H.; Rich, E.A. Multifaceted activities of the HIV-1 transactivator of transcription. Tat. J. Biol. Chem. 1999, 274, 28837–28840. [Google Scholar] [CrossRef]
- Marcello, A.; Zoppe, M.; Giacca, M. Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life 2001, 51, 175–181. [Google Scholar] [CrossRef]
- Zapp, M.L.; Green, M.R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature 1989, 342, 714–716. [Google Scholar]
- Kjems, J.; Brown, M.; Chang, D.D.; Sharp, P.A. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc. Natl. Acad. Sci. USA 1991, 88, 683–687. [Google Scholar]
- Chang, D.D.; Sharp, P.A. Regulation by HIV Rev depends upon recognition of splice sites. Cell 1989, 59, 789–795. [Google Scholar] [CrossRef]
- Cullen, B.R. Nuclear RNA export pathways. Mol. Cell Biol. 2000, 20, 4181–4187. [Google Scholar] [CrossRef]
- Hope, T.J. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 1999, 365, 186–191. [Google Scholar] [CrossRef]
- Pollard, V.W.; Malim, M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998, 52, 491–532. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Jeang, K.T. Rev-ing up post-transcriptional HIV-1 RNA expression. RNA Biol. 2011, 8, 195–199. [Google Scholar] [CrossRef]
- Schroder, A.R.; Shinn, P.; Chen, H.; Berry, C.; Ecker, J.R.; Bushman, F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002, 110, 521–529. [Google Scholar] [CrossRef]
- Dieudonne, M.; Maiuri, P.; Biancotto, C.; Knezevich, A.; Kula, A.; Lusic, M.; Marcello, A. Transcriptional competence of the integrated HIV-1 provirus at the nuclear periphery. EMBO J. 2009, 28, 2231–2243. [Google Scholar] [CrossRef]
- Marcello, A.; Dhir, S.; Dieudonne, M. Nuclear positional control of HIV transcription in 4D. Nucleus 2010, 1, 8–11. [Google Scholar]
- Maiuri, P.; Knezevich, A.; Bertrand, E.; Marcello, A. Real-time imaging of the HIV-1 transcription cycle in single living cells. Methods 2011, 53, 62–67. [Google Scholar] [CrossRef]
- Boireau, S.; Maiuri, P.; Basyuk, E.; de la Mata, M.; Knezevich, A.; Pradet-Balade, B.; Backer, V.; Kornblihtt, A.; Marcello, A.; Bertrand, E. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol. 2007, 179, 291–304. [Google Scholar] [CrossRef]
- Molle, D.; Maiuri, P.; Boireau, S.; Bertrand, E.; Knezevich, A.; Marcello, A.; Basyuk, E. A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites. Retrovirology 2007, 4, 36. [Google Scholar] [CrossRef]
- De Marco, A.; Biancotto, C.; Knezevich, A.; Maiuri, P.; Vardabasso, C.; Marcello, A. Intragenic transcriptional cis-activation of the human immunodeficiency virus 1 does not result in allele-specific inhibition of the endogenous gene. Retrovirology 2008, 5, 98. [Google Scholar] [CrossRef]
- Maiuri, P.; Knezevich, A.; De Marco, A.; Mazza, D.; Kula, A.; McNally, J.G.; Marcello, A. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 2011, 12, 1280–1285. [Google Scholar] [CrossRef]
- Marcello, A. RNA polymerase II transcription on the fast lane. Transcription 2012, 3, 29–34. [Google Scholar] [CrossRef]
- Kula, A.; Guerra, J.; Knezevich, A.; Kleva, D.; Myers, M.P.; Marcello, A. Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function. Retrovirology 2011, 8, 60. [Google Scholar] [CrossRef]
- Purcell, D.F.; Martin, M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar]
- Schwartz, S.; Felber, B.K.; Benko, D.M.; Fenyo, E.M.; Pavlakis, G.N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 1990, 64, 2519–2529. [Google Scholar]
- Staffa, A.; Cochrane, A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell Biol. 1995, 15, 4597–4605. [Google Scholar]
- Staffa, A.; Cochrane, A. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3' splice site. J. Virol. 1994, 68, 3071–3079. [Google Scholar]
- Dyhr-Mikkelsen, H.; Kjems, J. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J. Biol. Chem. 1995, 270, 24060–24066. [Google Scholar]
- O'Reilly, M.M.; McNally, M.T.; Beemon, K.L. Two strong 5' splice sites and competing, suboptimal 3' splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology 1995, 213, 373–385. [Google Scholar] [CrossRef]
- Si, Z.; Amendt, B.A.; Stoltzfus, C.M. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3' splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997, 25, 861–867. [Google Scholar] [CrossRef]
- Amendt, B.A.; Si, Z.H.; Stoltzfus, C.M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: Evidence for inhibition mediated by cellular factors. Mol. Cell Biol. 1995, 15, 4606–4615. [Google Scholar]
- Amendt, B.A.; Hesslein, D.; Chang, L.J.; Stoltzfus, C.M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell Biol. 1994, 14, 3960–3970. [Google Scholar]
- Caputi, M.; Mayeda, A.; Krainer, A.R.; Zahler, A.M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. Embo. J. 1999, 18, 4060–4067. [Google Scholar] [CrossRef]
- Tange, T.O.; Damgaard, C.K.; Guth, S.; Valcarcel, J.; Kjems, J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. Embo. J. 2001, 20, 5748–5758. [Google Scholar] [CrossRef]
- Cochrane, A.W.; Jones, K.S.; Beidas, S.; Dillon, P.J.; Skalka, A.M.; Rosen, C.A. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J. Virol. 1991, 65, 5305–5313. [Google Scholar]
- Maldarelli, F.; Martin, M.A.; Strebel, K. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: Novel level of gene regulation. J. Virol. 1991, 65, 5732–5743. [Google Scholar]
- Nasioulas, G.; Zolotukhin, A.S.; Tabernero, C.; Solomin, L.; Cunningham, C.P.; Pavlakis, G.N.; Felber, B.K. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J. Virol. 1994, 68, 2986–2993. [Google Scholar]
- Schneider, R.; Campbell, M.; Nasioulas, G.; Felber, B.K.; Pavlakis, G.N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 1997, 71, 4892–4903. [Google Scholar]
- Schwartz, S.; Campbell, M.; Nasioulas, G.; Harrison, J.; Felber, B.K.; Pavlakis, G.N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 1992, 66, 7176–7182. [Google Scholar]
- Mikaelian, I.; Krieg, M.; Gait, M.J.; Karn, J. Interactions of INS (CRS) elements and the splicing machinery regulate the production of Rev-responsive mRNAs. J. Mol. Biol. 1996, 257, 246–264. [Google Scholar] [CrossRef]
- Najera, I.; Krieg, M.; Karn, J. Synergistic stimulation of HIV-1 rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1. J. Mol. Biol. 1999, 285, 1951–1964. [Google Scholar] [CrossRef]
- Afonina, E.; Neumann, M.; Pavlakis, G.N. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 1997, 272, 2307–2311. [Google Scholar]
- Black, A.C.; Luo, J.; Chun, S.; Bakker, A.; Fraser, J.K.; Rosenblatt, J.D. Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes 1996, 12, 275–285. [Google Scholar]
- Zolotukhin, A.S.; Michalowski, D.; Bear, J.; Smulevitch, S.V.; Traish, A.M.; Peng, R.; Patton, J.; Shatsky, I.N.; Felber, B.K. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell Biol. 2003, 23, 6618–6630. [Google Scholar]
- Sodroski, J.; Goh, W.C.; Rosen, C.; Dayton, A.; Terwilliger, E.; Haseltine, W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 1986, 321, 412–417. [Google Scholar]
- Feinberg, M.B.; Jarrett, R.F.; Aldovini, A.; Gallo, R.C.; Wong-Staal, F. HTLV-II expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell 1986, 46, 807–817. [Google Scholar] [CrossRef]
- Meggio, F.; D'Agostino, D.M.; Ciminale, V.; Chieco-Bianchi, L.; Pinna, L.A. Phosphorylation of HIV-1 Rev protein: Implication of protein kinase CK2 and pro-directed kinases. Biochem Biophys Res Commun 1996, 226, 547–554. [Google Scholar] [CrossRef]
- Invernizzi, C.F.; Xie, B.; Richard, S.; Wainberg, M.A. PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA. Retrovirology 2006, 3, 93. [Google Scholar] [CrossRef]
- Groom, H.C.; Anderson, E.C.; Lever, A.M. Rev: Beyond nuclear export. J. Gen. Virol. 2009, 90, 1303–1318. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Jeang, K.T. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV- RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 14787–14792. [Google Scholar] [CrossRef]
- Grewe, B.; Uberla, K. The human immunodeficiency virus type 1 Rev protein: Menage a trois during the early phase of the lentiviral replication cycle. J. Gen. Virol. 2010, 91, 1893–1897. [Google Scholar] [CrossRef]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Henderson, B.R.; Percipalle, P. Interactions between HIV Rev and nuclear import and export factors: The Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 1997, 274, 693–707. [Google Scholar] [CrossRef]
- Truant, R.; Cullen, B.R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell Biol. 1999, 19, 1210–1217. [Google Scholar]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef]
- Fang, J.; Acheampong, E.; Dave, R.; Wang, F.; Mukhtar, M.; Pomerantz, R.J. The RNA helicase DDX1 is involved in restricted HIV-1 Rev function in human astrocytes. Virology 2005, 336, 299–307. [Google Scholar] [CrossRef]
- Li, J.; Tang, H.; Mullen, T.M.; Westberg, C.; Reddy, T.R.; Rose, D.W.; Wong-Staal, F. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc. Natl. Acad. Sci. USA 1999, 96, 709–714. [Google Scholar]
- Ma, J.; Rong, L.; Zhou, Y.; Roy, B.B.; Lu, J.; Abrahamyan, L.; Mouland, A.J.; Pan, Q.; Liang, C. The requirement of the DEAD-box protein DDX24 for the packaging of human immunodeficiency virus type 1 RNA. Virology 2008, 375, 253–264. [Google Scholar] [CrossRef]
- Fankhauser, C.; Izaurralde, E.; Adachi, Y.; Wingfield, P.; Laemmli, U.K. Specific complex of human immunodeficiency virus type 1 rev and nucleolar B23 proteins: Dissociation by the Rev response element. Mol. Cell Biol. 1991, 11, 2567–2575. [Google Scholar]
- Tange, T.O.; Jensen, T.H.; Kjems, J. In vitro interaction between human immunodeficiency virus type 1 Rev protein and splicing factor ASF/SF2-associated protein, p32. J. Biol. Chem. 1996, 271, 10066–10072. [Google Scholar]
- Cochrane, A.; Murley, L.L.; Gao, M.; Wong, R.; Clayton, K.; Brufatto, N.; Canadien, V.; Mamelak, D.; Chen, T.; Richards, D.; Zeghouf, M.; Greenblatt, J.; Burks, C.; Frappier, L. Stable complex formation between HIV Rev and the nucleosome assembly protein, NAP1, affects Rev function. Virology 2009, 388, 103–111. [Google Scholar] [CrossRef]
- Gu, L.; Tsuji, T.; Jarboui, M.A.; Yeo, G.P.; Sheehy, N.; Hall, W.W.; Gautier, V.W. Intermolecular masking of the HIV-1 Rev NLS by the cellular protein HIC: Novel insights into the regulation of Rev nuclear import. Retrovirology 2011, 8, 17. [Google Scholar] [CrossRef]
- Ruhl, M.; Himmelspach, M.; Bahr, G.M.; Hammerschmid, F.; Jaksche, H.; Wolff, B.; Aschauer, H.; Farrington, G.K.; Probst, H.; Bevec, D.; et al. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J. Cell Biol. 1993, 123, 1309–1320. [Google Scholar] [CrossRef]
- Bevec, D.; Jaksche, H.; Oft, M.; Wohl, T.; Himmelspach, M.; Pacher, A.; Schebesta, M.; Koettnitz, K.; Dobrovnik, M.; Csonga, R.; Lottspeich, F.; Hauber, J. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science 1996, 271, 1858–1860. [Google Scholar]
- Campbell, L.H.; Borg, K.T.; Haines, J.K.; Moon, R.T.; Schoenberg, D.R.; Arrigo, S.J. Human immunodeficiency virus type 1 Rev is required in vivo for binding of poly(A)-binding protein to Rev-dependent RNAs. J. Virol. 1994, 68, 5433–5438. [Google Scholar]
- Bogerd, H.P.; Fridell, R.A.; Madore, S.; Cullen, B.R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell 1995, 82, 485–494. [Google Scholar] [CrossRef]
- Fritz, C.C.; Zapp, M.L.; Green, M.R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature 1995, 376, 530–533. [Google Scholar]
- Reddy, T.R.; Xu, W.; Mau, J.K.; Goodwin, C.D.; Suhasini, M.; Tang, H.; Frimpong, K.; Rose, D.W.; Wong-Staal, F. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat. Med. 1999, 5, 635–642. [Google Scholar] [CrossRef]
- Modem, S.; Badri, K.R.; Holland, T.C.; Reddy, T.R. Sam68 is absolutely required for Rev function and HIV-1 production. Nucleic Acids Res. 2005, 33, 873–879. [Google Scholar] [CrossRef]
- Modem, S.; Reddy, T.R. An anti-apoptotic protein, Hax-1, inhibits the HIV-1 rev function by altering its sub-cellular localization. J. Cell Physiol. 2008, 214, 14–19. [Google Scholar]
- Yedavalli, V.S.; Jeang, K.T. Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression. Retrovirology 2011, 8, 61. [Google Scholar] [CrossRef]
- Powell, D.M.; Amaral, M.C.; Wu, J.Y.; Maniatis, T.; Greene, W.C. HIV Rev-dependent binding of SF2/ASF to the Rev response element: possible role in Rev-mediated inhibition of HIV RNA splicing. Proc. Natl. Acad. Sci. USA 1997, 94, 973–978. [Google Scholar]
- Xu, Y.; Reddy, T.R.; Fischer, W.H.; Wong-Staal, F. A Novel hnRNP Specifically Interacts with HIV-1 RRE RNA. J. Biomed. Sci. 1996, 3, 82–91. [Google Scholar] [CrossRef]
- Kaminski, R.; Darbinian, N.; Sawaya, B.E.; Slonina, D.; Amini, S.; Johnson, E.M.; Rappaport, J.; Khalili, K.; Darbinyan, A. Puralpha as a cellular co-factor of Rev/RRE-mediated expression of HIV-1 intron-containing mRNA. J. Cell Biochem. 2008, 103, 1231–1245. [Google Scholar] [CrossRef]
- Kubota, S.; Adachi, Y.; Copeland, T.D.; Oroszlan, S. Binding of human prothymosin alpha to the leucine-motif/activation domains of HTLV-I Rex and HIV-1 Rev. Eur. J. Biochem. 1995, 233, 48–54. [Google Scholar]
- Urcuqui-Inchima, S.; Castano, M.E.; Hernandez-Verdun, D.; St-Laurent, G., 3rd; Kumar, A. Nuclear Factor 90, a cellular dsRNA binding protein inhibits the HIV Rev-export function. Retrovirology 2006, 3, 83. [Google Scholar] [CrossRef]
- Wu, B.Y.; Woffendin, C.; Duckett, C.S.; Ohno, T.; Nabel, G.J. Regulation of human retroviral latency by the NF-kappa B/I kappa B family: Inhibition of human immunodeficiency virus replication by I kappa B through a Rev-dependent mechanism. Proc. Natl. Acad. Sci. USA 1995, 92, 1480–1484. [Google Scholar]
- Kramer-Hammerle, S.; Ceccherini-Silberstein, F.; Bickel, C.; Wolff, H.; Vincendeau, M.; Werner, T.; Erfle, V.; Brack-Werner, R. Identification of a novel Rev-interacting cellular protein. BMC Cell Biol. 2005, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Ariumi, Y.; Trono, D. Ataxia-telangiectasia-mutated (ATM) protein can enhance human immunodeficiency virus type 1 replication by stimulating Rev function. J. Virol. 2006, 80, 2445–2452. [Google Scholar] [CrossRef]
- Kimura, T.; Hashimoto, I.; Nishikawa, M.; Fujisawa, J.I. A role for Rev in the association of HIV-1 gag mRNA with cytoskeletal beta-actin and viral protein expression. Biochimie 1996, 78, 1075–1080. [Google Scholar] [CrossRef]
- Hofmann, W.; Reichart, B.; Ewald, A.; Muller, E.; Schmitt, I.; Stauber, R.H.; Lottspeich, F.; Jockusch, B.M.; Scheer, U.; Hauber, J.; Dabauvalle, M.C. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 2001, 152, 895–910. [Google Scholar] [CrossRef]
- Askjaer, P.; Jensen, T.H.; Nilsson, J.; Englmeier, L.; Kjems, J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 1998, 273, 33414–33422. [Google Scholar]
- Brennan, C.M.; Gallouzi, I.E.; Steitz, J.A. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 2000, 151, 1–14. [Google Scholar]
- Jeang, K.T.; Yedavalli, V. Role of RNA helicases in HIV-1 replication. Nucleic Acids Res. 2006, 34, 4198–4205. [Google Scholar] [CrossRef]
- Fang, J.; Kubota, S.; Yang, B.; Zhou, N.; Zhang, H.; Godbout, R.; Pomerantz, R.J. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology 2004, 330, 471–480. [Google Scholar] [CrossRef]
- Dundr, M.; Leno, G.H.; Hammarskjold, M.L.; Rekosh, D.; Helga-Maria, C.; Olson, M.O. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J. Cell Sci. 1995, 108, 2811–2823. [Google Scholar]
- Zheng, Y.H.; Yu, H.F.; Peterlin, B.M. Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell Biol. 2003, 5, 611–618. [Google Scholar] [CrossRef]
- Li, Y.P. Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J. Virol. 1997, 71, 4098–4102. [Google Scholar]
- Gautier, V.W.; Sheehy, N.; Duffy, M.; Hashimoto, K.; Hall, W.W. Direct interaction of the human I-mfa domain-containing protein, HIC, with HIV-1 Tat results in cytoplasmic sequestration and control of Tat activity. Proc. Natl. Acad. Sci. USA 2005, 102, 16362–16367. [Google Scholar]
- Berro, R.; Kehn, K.; de la Fuente, C.; Pumfery, A.; Adair, R.; Wade, J.; Colberg-Poley, A.M.; Hiscott, J.; Kashanchi, F. Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32. J. Virol. 2006, 80, 3189–3204. [Google Scholar]
- Vardabasso, C.; Manganaro, L.; Lusic, M.; Marcello, A.; Giacca, M. The histone chaperone protein Nucleosome Assembly Protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology 2008, 5, 8. [Google Scholar] [CrossRef]
- De Marco, A.; Dans, P.D.; Knezevich, A.; Maiuri, P.; Pantano, S.; Marcello, A. Subcellular localization of the interaction between the human immunodeficiency virus transactivator Tat and the nucleosome assembly protein 1. Amino Acids 2010, 38, 1583–1593. [Google Scholar] [CrossRef]
- Sanchez-Velar, N.; Udofia, E.B.; Yu, Z.; Zapp, M.L. hRIP, a cellular cofactor for Rev function, promotes release of HIV RNAs from the perinuclear region. Genes Dev. 2004, 18, 23–34. [Google Scholar] [CrossRef]
- Yu, Z.; Sanchez-Velar, N.; Catrina, I.E.; Kittler, E.L.; Udofia, E.B.; Zapp, M.L. The cellular HIV-1 Rev cofactor hRIP is required for viral replication. Proc. Natl. Acad. Sci. USA 2005, 102, 4027–4032. [Google Scholar]
- Zhang, Z.; Carmichael, G.G. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 2001, 106, 465–475. [Google Scholar] [CrossRef]
- Cronshaw, J.M.; Krutchinsky, A.N.; Zhang, W.; Chait, B.T.; Matunis, M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002, 158, 915–927. [Google Scholar] [CrossRef]
- Naji, S.; Ambrus, G.; Cimermancic, P.; Reyes, J.R.; Johnson, J.R.; Filbrandt, R.; Huber, M.D.; Vesely, P.; Krogan, N.J.; Yates, J.R.; Saphire, A.C.; Gerace, L. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell Prot. 2012, 11, M111–015313. [Google Scholar] [CrossRef]
- Jager, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; Hernandez, H.; Jang, G.M.; Roth, S.L.; Akiva, E.; Marlett, J.; Stephens, M.; D'Orso, I.; Fernandes, J.; Fahey, M.; Mahon, C.; O'Donoghue, A.J.; Todorovic, A.; Morris, J.H.; Maltby, D.A.; Alber, T.; Cagney, G.; Bushman, F.D.; Young, J.A.; Chanda, S.K.; Sundquist, W.I.; Kortemme, T.; Hernandez, R.D.; Craik, C.S.; Burlingame, A.; Sali, A.; Frankel, A.D.; Krogan, N.J. Global landscape of HIV-human protein complexes. Nature 2012, 481, 365–370. [Google Scholar]
- Iacampo, S.; Cochrane, A. Human immunodeficiency virus type 1 Rev function requires continued synthesis of its target mRNA. J. Virol. 1996, 70, 8332–8339. [Google Scholar]
- Luo, Y.; Madore, S.J.; Parslow, T.G.; Cullen, B.R.; Peterlin, B.M. Functional analysis of interactions between Tat and the trans-activation response element of human immunodeficiency virus type 1 in cells. J. Virol. 1993, 67, 5617–5622. [Google Scholar]
- Southgate, C.; Zapp, M.L.; Green, M.R. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature 1990, 345, 640–642. [Google Scholar]
- Berthold, E.; Maldarelli, F. cis-acting elements in human immunodeficiency virus type 1 RNAs direct viral transcripts to distinct intranuclear locations. J. Virol. 1996, 70, 4667–4682. [Google Scholar]
- Kalland, K.H.; Szilvay, A.M.; Langhoff, E.; Haukenes, G. Subcellular distribution of human immunodeficiency virus type 1 Rev and colocalization of Rev with RNA splicing factors in a speckled pattern in the nucleoplasm. J. Virol. 1994, 68, 1475–1485. [Google Scholar]
- Wolff, B.; Cohen, G.; Hauber, J.; Meshcheryakova, D.; Rabeck, C. Nucleocytoplasmic transport of the Rev protein of human immunodeficiency virus type 1 is dependent on the activation domain of the protein. Exp. Cell Res. 1995, 217, 31–41. [Google Scholar] [CrossRef]
- Boe, S.O.; Bjorndal, B.; Rosok, B.; Szilvay, A.M.; Kalland, K.H. Subcellular localization of human immunodeficiency virus type 1 RNAs, Rev, and the splicing factor SC-35. Virology 1998, 244, 473–482. [Google Scholar] [CrossRef]
- Favaro, J.P.; Borg, K.T.; Arrigo, S.J.; Schmidt, M.G. Effect of Rev on the intranuclear localization of HIV-1 unspliced RNA. Virology 1998, 249, 286–296. [Google Scholar] [CrossRef]
- Zhang, G.; Zapp, M.L.; Yan, G.; Green, M.R. Localization of HIV-1 RNA in mammalian nuclei. J. Cell Biol. 1996, 135, 9–18. [Google Scholar] [CrossRef]
- Pond, S.J.; Ridgeway, W.K.; Robertson, R.; Wang, J.; Millar, D.P. HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc. Natl. Acad. Sci. USA 2009, 106, 1404–1408. [Google Scholar]
- Cook, K.S.; Fisk, G.J.; Hauber, J.; Usman, N.; Daly, T.J.; Rusche, J.R. Characterization of HIV-1 REV protein: Binding stoichiometry and minimal RNA substrate. Nucleic Acids Res 1991, 19, 1577–1583. [Google Scholar]
- Daly, T.J.; Cook, K.S.; Gray, G.S.; Maione, T.E.; Rusche, J.R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 1989, 342, 816–819. [Google Scholar]
- Heaphy, S.; Dingwall, C.; Ernberg, I.; Gait, M.J.; Green, S.M.; Karn, J.; Lowe, A.D.; Singh, M.; Skinner, M.A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell 1990, 60, 685–693. [Google Scholar] [CrossRef]
- Malim, M.H.; Tiley, L.S.; McCarn, D.F.; Rusche, J.R.; Hauber, J.; Cullen, B.R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 1990, 60, 675–683. [Google Scholar] [CrossRef]
- Madore, S.J.; Tiley, L.S.; Malim, M.H.; Cullen, B.R. Sequence requirements for Rev multimerization in vivo. Virology 1994, 202, 186–194. [Google Scholar] [CrossRef]
- Daugherty, M.D.; Liu, B.; Frankel, A.D. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010, 17, 1337–1342. [Google Scholar]
- Daugherty, M.D.; Booth, D.S.; Jayaraman, B.; Cheng, Y.; Frankel, A.D. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Natl. Acad. Sci. USA 2010, 107, 12481–12486. [Google Scholar]
- Daugherty, M.D.; D'Orso, I.; Frankel, A.D. A solution to limited genomic capacity: Using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol. Cell 2008, 31, 824–834. [Google Scholar] [CrossRef]
- DiMattia, M.A.; Watts, N.R.; Stahl, S.J.; Rader, C.; Wingfield, P.T.; Stuart, D.I.; Steven, A.C.; Grimes, J.M. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc. Natl. Acad. Sci. USA 2010, 107, 5810–5814. [Google Scholar]
- Hoffmann, D.; Schwark, D.; Banning, C.; Brennen, M.; Lakshmikanth, M.; Krepstakies, M.; Schindler, M.; Millar, D.P.; Hauber, J. Formation of trans-activation competent HIV-1 Rev: RRE complexes requires the recruitment of multiple protein activation domains. PLoS One 2012, in press. [Google Scholar]
- Robertson-Anderson, R.M.; Wang, J.; Edgcomb, S.P.; Carmel, A.B.; Williamson, J.R.; Millar, D.P. Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element. J. Mol. Biol. 2011, 410, 959–971. [Google Scholar] [CrossRef]
- Grunwald, D.; Singer, R.H. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature 2010, 467, 604–607. [Google Scholar]
- Mor, A.; Shav-Tal, Y. Dynamics and kinetics of nucleo-cytoplasmic mRNA export. Wiley Interdiscip Rev. RNA 2010, 1, 388–401. [Google Scholar] [CrossRef]
- Müller, B. Novel imaging technologies in the study of HIV. Future Virol. 2011, 6, 929–940. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kula, A.; Marcello, A. Dynamic Post-Transcriptional Regulation of HIV-1 Gene Expression. Biology 2012, 1, 116-133. https://doi.org/10.3390/biology1020116
Kula A, Marcello A. Dynamic Post-Transcriptional Regulation of HIV-1 Gene Expression. Biology. 2012; 1(2):116-133. https://doi.org/10.3390/biology1020116
Chicago/Turabian StyleKula, Anna, and Alessandro Marcello. 2012. "Dynamic Post-Transcriptional Regulation of HIV-1 Gene Expression" Biology 1, no. 2: 116-133. https://doi.org/10.3390/biology1020116




