Psychrophily and Catalysis
Abstract
:1. Introduction
2. General Properties of Cold-Adapted Enzymes
- (1).
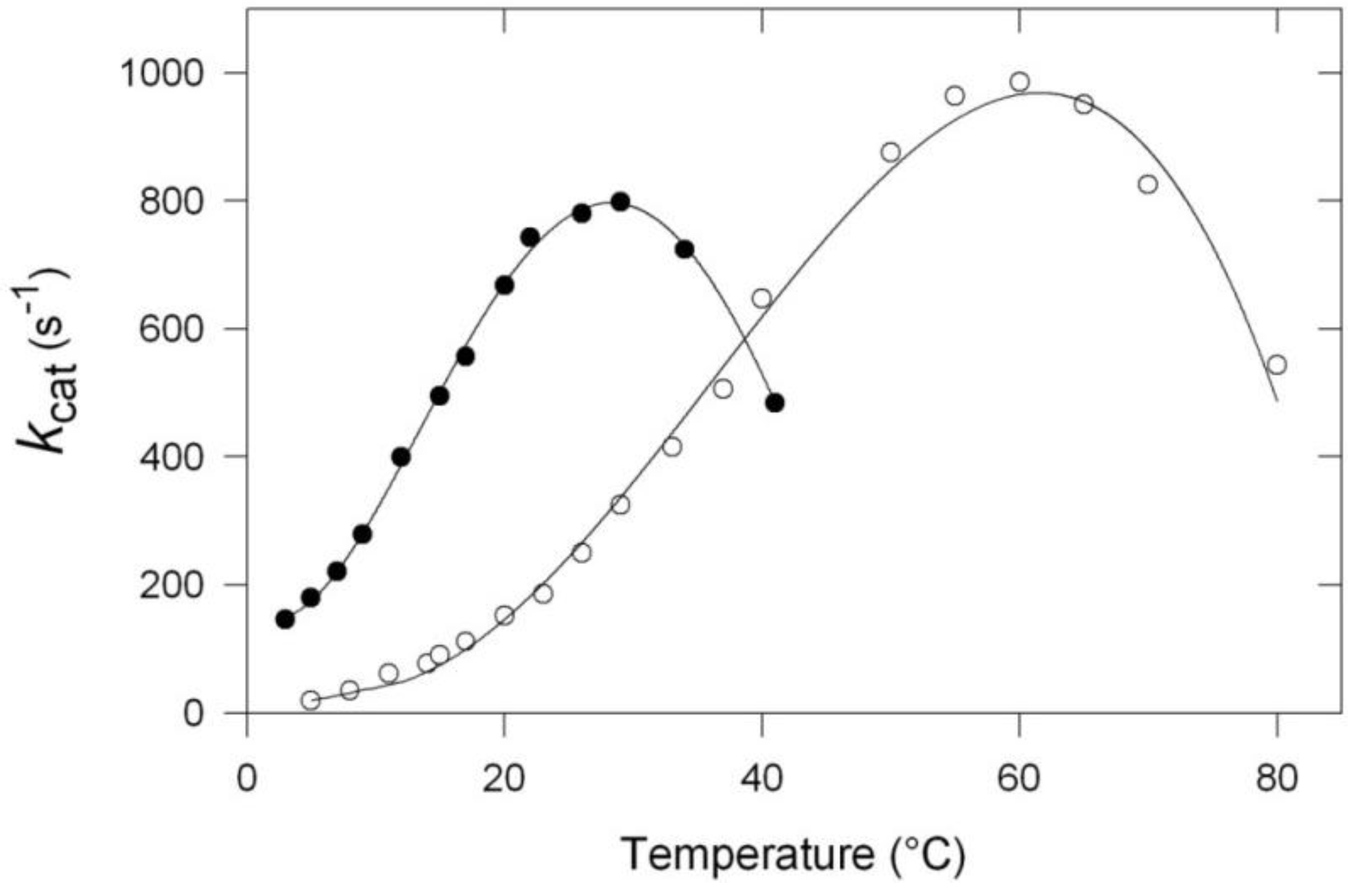
- The apparent optimum temperature of the cold-adapted enzyme is displaced towards low temperatures by as much as 30 °C.
- (2).
- The cold-adapted enzyme displays a much higher catalytic efficiency than the thermophilic enzyme up to approximately its apparent optimum.
- (3).
- The cold-adapted enzyme is, in contrast with the thermophilic one, rapidly inactivated at temperatures above 25 °C.
| Enzyme-organism | T (°C) | Km | Ref |
|---|---|---|---|
| Alpha-amylase | |||
| P: Pseudoalteromonas haloplanktis | 25 | 234.00 uM | [17] |
| M: Pig pancreatic | 25 | 65.00 | |
| Aspartate aminotransferase | |||
| P: P. haloplanktis | 07 | 5.82 mM | [18] |
| 25 | 8.34 | ||
| M: E.coli | 25 | 7.31 | |
| 35 | 21.04 | ||
| Aspartate transcarbamylase | |||
| P: Gram-TAD1 | 11 | 20.00 mM | [19] |
| M: E.coli | 30 | 0.014 | |
| Citrate synthase | |||
| P: Antarctic bacterium DS2-3R | 23 | 230 uM | [20] |
| M: mesophiles | <50 uM | ||
| DNA ligase | |||
| P: Pseudoalteromonas haloplanktis | 04 | 0.165 uM | [21] |
| M: E.coli | 18 | 0.179 | |
| T: Thermus scotoductus | 18 | 0.179 | |
| 30 | 0.702 | ||
| T: Thermus scotoductus | 45 | 0.236 | |
| Elongation factor TU | |||
| P: Moraxella TAC II 25 | 15 | 0.36 uM | [22] |
| M: E.coli | 15 | 0.13 | |
| Endonuclease I | |||
| P: Vibrio salmonicido | 05 | 246.00 mM | [23] |
| M: Vibrio cholerae | 05 | 118.00 | |
| Isocitrate dehydrogenase | |||
| P: Colwellia maris | 15 | 62.00 mM | [24] |
| M: E. coli | 15 | 3.30 | |
| Lactate dehydrogenase | |||
| P: Champsocephalus gunnarii | 00 | 0.16 mM | [25] |
| M: Deinococcus radiodurans | 48 | 0.21 | |
| T: Thermus thermophilus | 90 | 0.16 | |
| Ornithine transcarbamylase | |||
| P: Moritella abyssi | 05 | 1.78 mM | [26] |
| M: E. coli | 37 | 2.40 | |
| T: Thermus thermophilus | 55 | 0.10 | |
| RNA-dependent ATPase | |||
| P: Pseudoalteromonas haloplanktis | 10 | 0.60 mM | [27] |
| 25 | 0.9 | ||
| M: E.coli | 10 | 0.02 | |
| 25 | 0.06 | ||
| Subtilisin | |||
| P: Bacillus (Antarctic) | 05 | 26.00 uM | [28] |
| 25 | 37.00 | ||
| M: Bacillus licheniformis | 05 | 6.00 | |
| 25 | 17.00 | ||
| Triose phosphate isomerase | |||
| P: Vibrio marinus | 10 | 1.90 mM | [29] |
| M: E. coli | 25 | 1.09 | |
3. Activity and Stability
| Enzyme | Type | T (°C) | kcat(s−1) | ΔG* | ΔH* | TΔS* | Reference |
|---|---|---|---|---|---|---|---|
| kJ/mole | |||||||
| Amylase | Psy | 10 | 294.0 | 57.7 | 34.7 | −23.0 | [32] |
| Mes | 97.0 | 58.5 | 46.4 | −12.1 | |||
| Arginine kinase | Psy | 25 | 3.3 | 69.4 | 18.8 | −50.6 | [33] |
| Mes | 13.4 | 66.6 | 41.9 | −4.7 | |||
| Cellulase | Psy | 4 | 0.18 | 71.6 | 46.2 | −25.4 | [34] |
| Mes | 0.01 | 78.2 | 65.8 | −12.4 | |||
| Chitinase | Psy | 15 | 1.7 | 69.2 | 60.2 | −9.0 | [31] |
| Mes | 3.9 | 67.2 | 74.3 | +7.1 | |||
| Chitobiase | Psy | 15 | 3.8 | 59.5 | 44.7 | −14.8 | [35] |
| Mes | 0.9 | 63.5 | 71.5 | +8.0 | |||
| Citrate synthase | Psy | 7.4 | ΔS* = 22.7 | [20] | |||
| Mes | 11.5 | ΔS* = 9.7 | |||||
| Endonuclease | Psy | 5 | 9.41 | 62.8 | 33.4 | −29.4 | [23] |
| Mes | 1.03 | 67.9 | 74.0 | +6.1 | |||
| LDH | Psy | 0 | 250.0 | 75.0 | 22.0 | −53.0 | [25] |
| Mes | 72.0 | 75.0 | 45.0 | −30.0 | |||
| Lysozyme | Psy | 45.1 | 31.9 | −13.2 | [36] | ||
| Mes | 46.2 | 49.4 | +3.2 | ||||
| Subtilisin | Psy | 15 | 25.4 | 62.0 | 36.0 | −26.5 | [37] |
| Mes | 5.4 | 66.0 | 46.0 | −20.2 | |||
| Xylanase (bact) | Psy | 10 | 515.5 | 54.0 | 21.0 | −33.0 | [38] |
| Mes | 59.5 | 60.0 | 58.0 | −2.0 | |||
| Xylanase (yeast) | Psy | 5 | 14.8 | 52.3 | 45.3 | −7.0 | [39] |
| Mes | 4.9 | 54.6 | 49.9 | −4.7 | |||
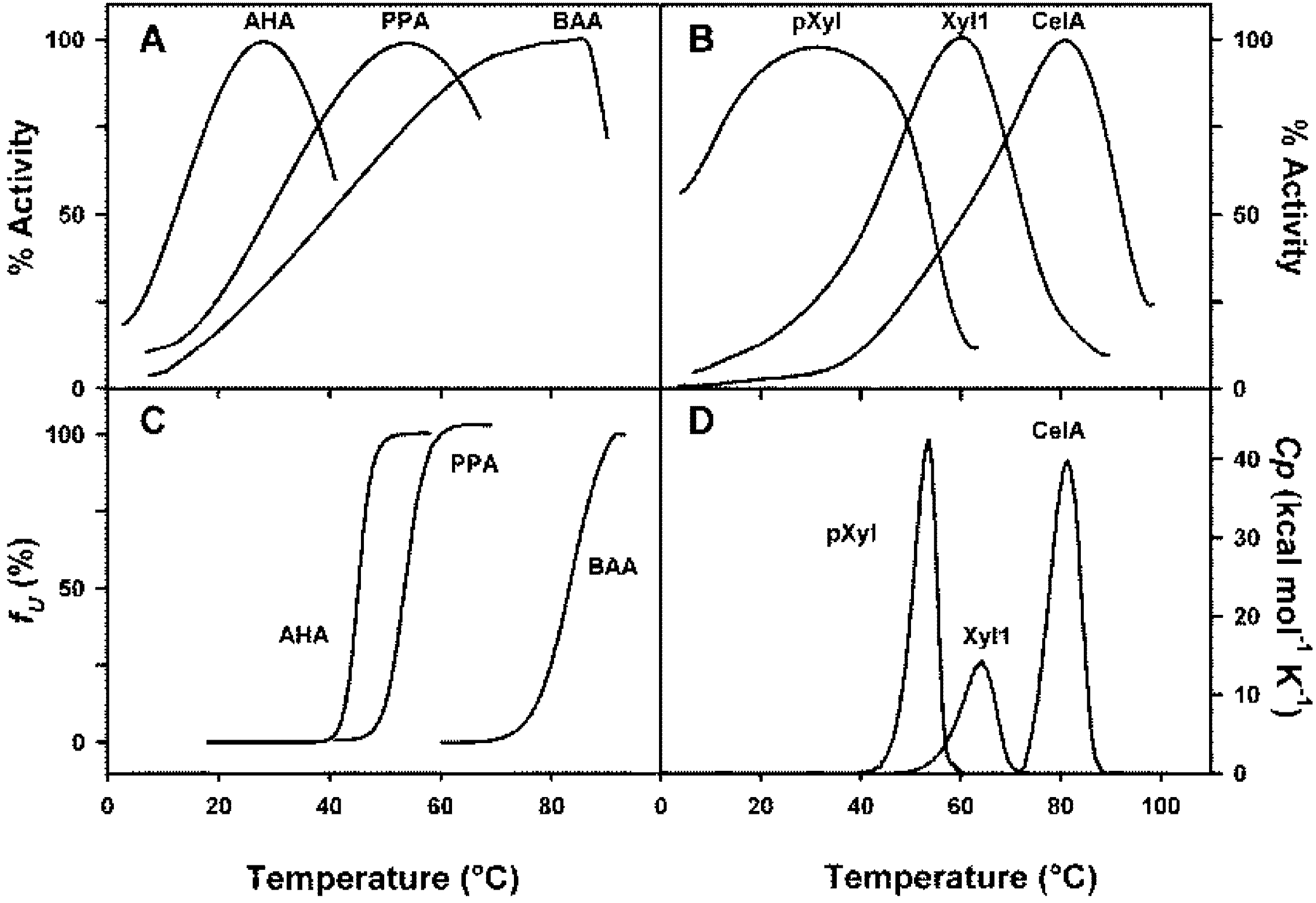
4. Thermodynamic Stability
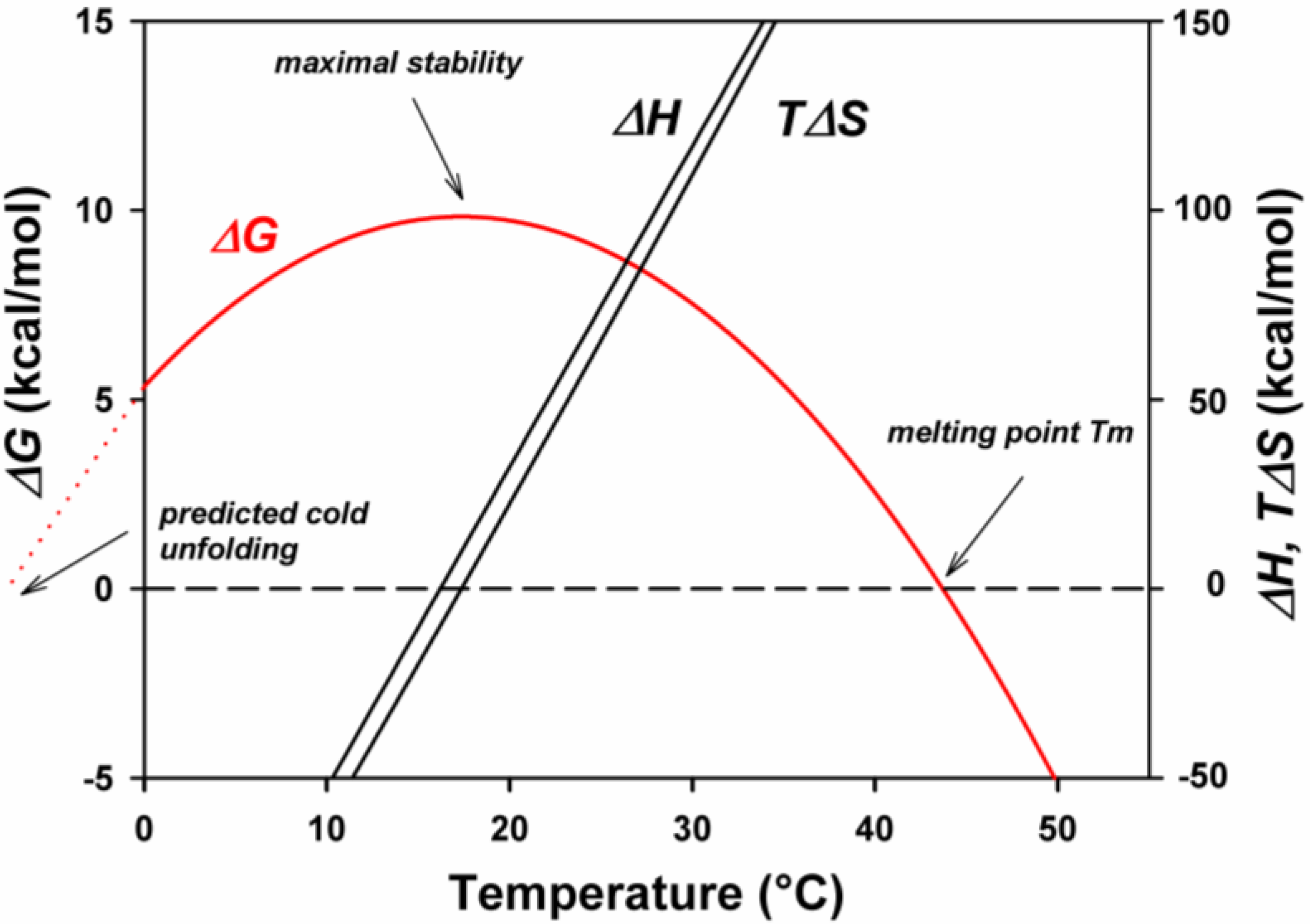
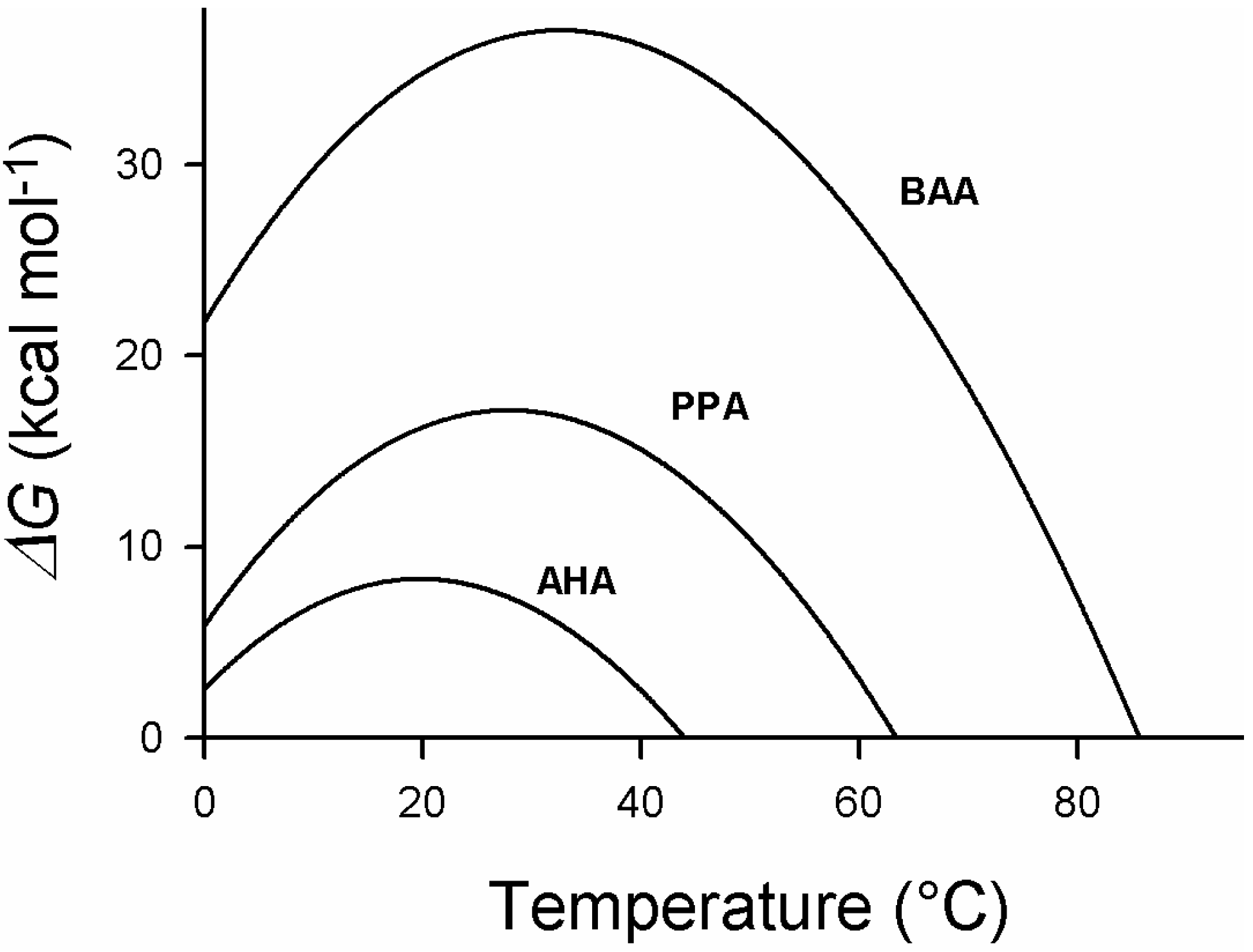
5. Engineering Cold-Adapted Enzymes
| Cold-Adapted Enzyme | Host Organism | Reference |
|---|---|---|
| Adenylate kinase | Bacillus globisporus | [52] |
| Adenylate kinase | Marinibacillus marinus | [53] |
| Alkaline metalloprotease | Pseudomonas sp. | [54] |
| Alkaline phosphatase | Pandalus borealis | [55] |
| Alkaline phosphatase | Bacterial strain TAB5 | [56] |
| Anionic trypsin | Salmo salar | [57] |
| Alpha-amylase | Pseudoalteromonas haloplanktis | [58] |
| Aspartate carbamoyltransferase | Moritella profunda | [59] |
| Beta-galactosidase | Arthrobacter sp. C2-2 | [60] |
| Catalase | Vibrio salmonicida | [61] |
| Cellulase | Pseudoalteromonas haloplanktis | [62] |
| Citrate synthase | Arthrobacter sp. strain DS2-3R | [63] |
| Elastase | Salmo salar | [64] |
| Endonuclease I | Vibrio salmonicida | [23] |
| Esterase | Pseudoalteromonas sp. 643A | [65] |
| Lactate dehydrogenase | Champsocephalus gunnari | [25] |
| Lipase B | Candida antarctica | [66] |
| Malate dehydrogenase | Aquaspirillium articum | [67] |
| Pepsin | Gadus morhua | [68] |
| Phenylalanine hydroxylase | Colwellia psychrerythtaea | [69] |
| Proteinase K-like | Serratia sp. | [70] |
| Protein-tyrosinephosphatase | Shewanella sp. | [71] |
| S-formylglutathione hydrolase | Pseudoalteromonas haloplanktis | [72] |
| Subtilisin-like protease | Vibrio sp. PA-44 | [73] |
| Subtilisin S41 | Bacillus sp. | [74] |
| Superoxide dismutase | Aliivibrio salmonicida | [75] |
| Triose phosphate isomerase | Vibrio marinus | [29] |
| Trypsin | Oncorhynchus ketav | [76] |
| Uracil-DNA N-glycosylase | Gadus morhua | [77] |
| Xylanase | Pseudoalteromonas sp. | [78] |
6. Folding at Low Temperature
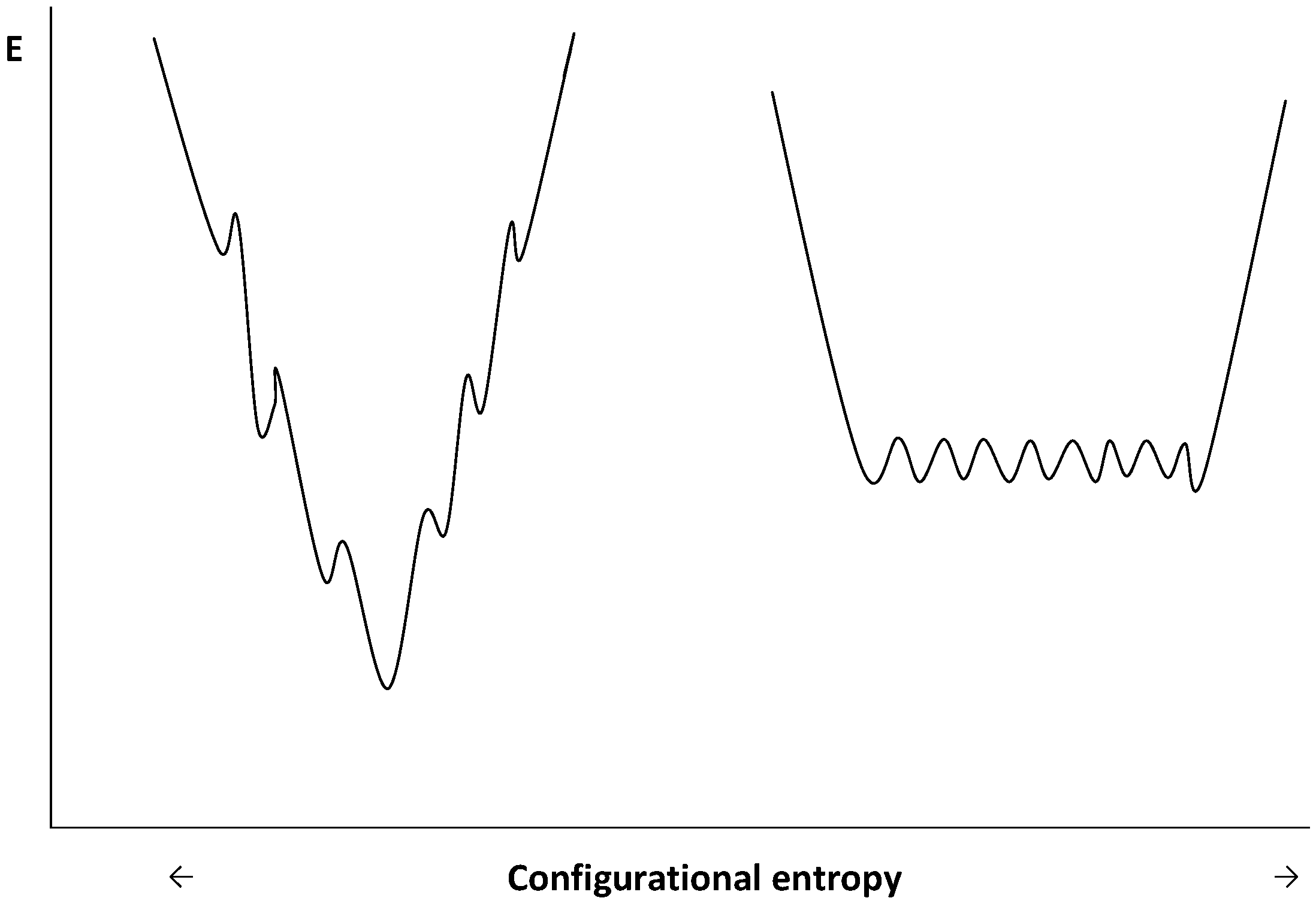
7. Conclusions
References
- Brinkmeyer, R.; Knittel, K.; Jürgens, J.; Weyland, H.; Amann, R.; Helmke, E. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl. Envirom. Microbiol. 2003, 69, 6610–6619. [Google Scholar] [CrossRef]
- Buckeridge, K.M.; Grogan, P. Deepened snow alters soil microbial nutrient limitations in arctic birch hummock tundra. Appl. Soil. Ecol. 2008, 39, 210–222. [Google Scholar] [CrossRef]
- Steven, B.; Briggs, S.; McKay, C.P.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Characterization of the microbial diversity in a permafrost sample from the Canadian high Arctic using culture-dependent and culture-independent methods. FEMS Microbiol. Ecol. 2007, 59, 513–523. [Google Scholar] [CrossRef]
- Shivagi, S.; Kumari, K.; Kishore, K.H.; Pindi, P.K.; Rao, P.S.; Srivinas, T.N.R.; Asthana, R.; Ravindra, R. Vertical distribution of bacteria in a lake sediment from Antarctica by culture-independent and culture-dependent approaches. Res. Microbiol. 2011, 162, 191–203. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Caruso, C.; Mangano, S.; Bruni, V.; De Domenico, M.; Michaud, L. Marine bacterioplankton diversity and community composition in an Antarctic coastal environment. Microbiol. Ecol. 2012, 63, 210–223. [Google Scholar] [CrossRef]
- Kirby, B.M.; Easton, S.; Tuffin, I.M.; Cowan, D.A. Bacterial diversity in polar habitats. In PolarMicrobiology. Life in a Deep Freeze; Miller, R.V., Whyte, L.G., Eds.; ASM Press: Washington DC, USA, 2012; Volume Chapter 1, p. 3. [Google Scholar]
- Collins, T.; D’Amico, S.; Marx, J.C.; Feller, G.; Gerday, C. Cold-adapted enzymes. In Physiology and Biochemistry of Extremophiles; Gerday, C.H., Glansdorff, N., Eds.; ASM Press: Washington DC, USA, 2007; Volume Chapter 13, p. 165. [Google Scholar]
- Garcia-Viloca, M.; Gao, J.; Karplus, M.; Truhlar, D.G. How enzymes work: Analysis by modern rate theory and computer simulation. Science 2004, 303, 186–195. [Google Scholar] [CrossRef]
- Demchenko, A.P.; Rusyn, O.I.; Saburova, E.A. Kinetics of the lactate dehydrogenase reaction in high viscosity media. Biochim. Biophys. Acta 1989, 998, 196–203. [Google Scholar] [CrossRef]
- Mastro, A.M.; Keith, A.D. Diffusion in the aqueous compartment. J. Cell Biol. 1984, 99, 180–187. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Bokhari, S.A.; Afzal, A.J.; Singh, S. A novel thermodynamic relationship based on Kramers theory for studying enzyme kinetics under high viscosity. IUBMB Life 2004, 56, 403–407. [Google Scholar] [CrossRef]
- Feller, G.; Lonhienne, T.; Deroanne, C.; Libioulle, C.; Van Beeumen, J.; Gerday, C. Purification, characterization, and nucleotide sequence of the thermolabile α-amylase from the Antarctic psychrotroph Alteromonas. haloplanktis A23. J. Biol. Chem. 1992, 267, 5217–5221. [Google Scholar]
- Kobori, H.; Sullivan, C.W.; Shizuya, H. Heat-labile alkaline phosphatase from Antarctic bacteria: Rapid 5' end-labeling of nucleic acid. Proc. Natl. Acad. Sci. USA 1984, 81, 6691–6695. [Google Scholar] [CrossRef]
- De Santi, C.; Tutino, M.L.; Mandrich, L.; Giuliani, M.; Parrilli, E.; Del Vecchio, P.; De Pascale, D. The hormone-sensitive lipase from Psychrobacter. sp. TA144: New insight in the structural/functional characterization. Biochimie 2010, 92, 949–957. [Google Scholar] [CrossRef]
- Simpson, P.J.L.; Codd, R. Cold adaptation of the mononuclear molybdoenzyme periplasmic nitrate reductase from the Antarctic bacterium Shewanella. gelidimarina. Biochem. Biophys. Res. Comm. 2011, 414, 783–788. [Google Scholar] [CrossRef]
- Angelaccio, S.; Florio, R.; Consalvi, V.; Festa, G.; Pascarella, P. Serine hydroxymethyltransferase from the cold adapted microorganism Psychromonas. ingrahamii: A low temperature active enzyme with broad specificity. Int. Mol. Sci. 2012, 13, 1314–1326. [Google Scholar] [CrossRef]
- D’Amico, S.; Gerday, C.; Feller, G. Structural determinants of cold adaptation and stability in a large protein. J. Biol. Chem. 2001, 276, 25791–25796. [Google Scholar] [CrossRef]
- Birolo, M.; Tutino, L. Aspartate aminotransferase from the Antarctic bacterium Pseudoalteromonas. haloplanktis TAC 125. Cloning, expression, properties, and molecular modelling. Eur. J. Biochem. 2000, 267, 2790–2802. [Google Scholar] [CrossRef]
- Sun, K.; Camardella, L.; Di Prisco, G.; Hervé, G. Properties of aspartate transcarbamylase from TAD1, a psychrophilic bacterial strain isolated from Antarctica. FEMS Microbiol. Lett. 1998, 164, 375–382. [Google Scholar] [CrossRef]
- Gerike, U.; Danson, M.J.; Russell, N.J.; Hough, D.W. Sequencing and expression of the gene encoding a cold-active citrate synthase from an Antarctic bacterium strain DS-3R. Eur. J. Biochem. 1997, 248, 49–57. [Google Scholar]
- Georlette, D.; Jonsson, Z.O.; Van Petegem, F.; Chessa, J.; Van Beeumen, J.; Hubscher, U.; Gerday, C. A DNA ligase from the psychrophile Pseudoalteromonas. haloplanktis gives insights into the adaptation of proteins to low temperatures. Eur. J. Biochem. 2000, 267, 3502–3512. [Google Scholar] [CrossRef] [Green Version]
- Masullo, M.; Arcari, P. Psychrophilic elongation factor TU from the Antarctic Moraxella. sp. TAC II 125: Biochemical characterization and cloning of the encoding gene. Biochemistry 2000, 39, 15531–15539. [Google Scholar] [CrossRef]
- Altermak, B.; Niiranen, L.; Willassen, N.P.; Smalas, A.O.; Moe, E. Comparative studies of endonuclease I from cold-adapted Vibrio. salmonicida and mesophilic Vibrio. cholerae. FEBS J. 2007, 274, 252–263. [Google Scholar] [CrossRef]
- Watanabe, S.; Yasutake, Y.; Tanaka, I.; Takada, Y. Elucidation of stability determinants of cold-adapted monomeric isocitrate dehydrogenase from a psychrophilic bazcterium, Colwellia. maris, by construction of chimeric enzymes. Microbiology 2005, 151, 1083–1094. [Google Scholar] [CrossRef]
- Coquelle, N.; Fioravanti, E.; Weik, M.; Vellieux, F.; Madern, D. Activity, stability and structural studies of lactate dehydrogenases adapted to extreme thermal environments. J. Mol. Biol. 2007, 374, 547–562. [Google Scholar] [CrossRef]
- Xu, Y.; Feller, G.; Gerday, C.; Glansdorff, N. Metabolic enzymes from psychrophilic bacteria: Challenge of adaptation to low temperatures in ornithine carbamoyltransferase from Moritella. abyssi. J. Bacteriol. 2003, 185, 2161–2168. [Google Scholar] [CrossRef]
- Cartier, G.; Lorieux, F.; Allemand, F.; Dreyfus, M.; Bizebard, T. Cold adaptation in DEAD-box proteins. Biochemistry 2010, 49, 2636–2646. [Google Scholar] [CrossRef]
- Narinx, E.; Baise, E.; Gerday, C. Subtilisin from antarctic bacteria: Characterization and site- directed mutagenesis of residues possibly involved in the adaptation to cold. Prot. Engineer. 1997, 10, 1271–1279. [Google Scholar] [CrossRef]
- Alvarez, M.; Zeelen, J.P.; Mainfroid, V.; Rentier-Delrue, F.; Martial, J.A.; Wyns, L. Triose-phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio. marinus. J. Biol. Chem. 1998, 273, 2199–2206. [Google Scholar]
- Hochachka, P.W.; Somero, G.N. Temperature. In Biochemical Adaptation; Hochachka, P.W., Domero, G.N., Eds.; Oxford University Press: New York, NY, USA, 2002; Volume Chapter 7, p. 290. [Google Scholar]
- Lonhienne, T.; Baise, E.; Feller, G.; Bouriotis, V.; Gerday, C. Enzyme activity determination on macromolecular substrates by isothermal titration calorimetry: Application to mesophilic and psychrophilic chitinases. Biochim. Biophys. Acta 2001, 1545, 349–356. [Google Scholar]
- D’Amico, S.; Marx, J.-C.; Gerday, C.; Feller, G. Activity-stability relationship in extremophilic enzymes. J. Biol. Chem. 2003, 278, 7891–7896. [Google Scholar]
- Suzuki, T.; Yamamoto, K.; Tada, H.; Uda, K. Cold-adapted features of arginine kinase from the deep-sea. Calyptogena. kaikoi. Mar. Biotechnol. 2012, 14, 294–303. [Google Scholar] [CrossRef]
- Garsoux, G.; Lamotte-Brasseur, J.; Gerday, C.; Feller, G. Kinetic and structural optimisation to catalysis at low temperatures in a psychrophilic cellulase from the Antarctic bacterium Pseudoalteromonas. haloplanktis. Biochem. J. 2004, 384, 247–253. [Google Scholar] [CrossRef]
- Lonhienne, T.; Zoidakis, J.; Vorgias, E.; Feller, G.; Gerday, C.; Bouriotis, V. Modular structure, local flexibility and cold-activity of a novel chitobiase from a psychrophilic Antarctic bacterium. J. Mol. Biol. 2001, 310, 291–297. [Google Scholar] [CrossRef]
- Sotelo-Mundo, R.R.; Lopez-Zavala, A.A.; Garcia-Orozco, K.D.; Arvizu-Flores, A.A.; Velazquez-Conteras, E.F.; Vaalenzuela-Soto, E.M.; Rojo-Dominguez, A.; Kanost, M.R. The lysozyme from insect (Manduca. sexta) is a cold-adapted enzyme. Protein Pept. Lett. 2007, 14, 774–778. [Google Scholar] [CrossRef]
- Davail, S.; Feller, G.; Narinx, E.; Gerday, C. Cold adaptation of proteins. Purification, characterization and sequence of the heat labile subtilisin from the Antarctic psychrophile Bacillus TA41. J. Biol. Chem. 1994, 269, 17448–17453. [Google Scholar]
- Collins, T.; Meuwis M-A. Gerday, C.; Feller, G. Activity, stability and flexibility in glycosidases adapted to extreme thermal environments. J. Mol. Biol. 2003, 328, 419–428. [Google Scholar] [CrossRef]
- Petrescu, J.; Lamotte-Brasseur, J.; Chessa, J.-P.; Ntarima, P.; Claeyssens, M.; Devreese, B.; Marino, G.; Gerday, C. Xylanase from psychrophilic yeast Cryptococcus. adeliae. Extremophiles 2000, 4, 137–144. [Google Scholar] [CrossRef]
- Marx, J.-C.; Collins, S.; D’Amico, S.; Feller, G.; Gerday, C. Cold-adapted enzymes from marine Antarctic microorganisms. Mar. Biotechnol. 2006, 9, 293–304. [Google Scholar]
- Robindon, G.W.; Cho, C.H. Role of hydration water in protein unfolding. Biophys. J. 1999, 77, 3331–3318. [Google Scholar]
- Loladze, V.V.; Ermolenko, D.N.; Makhatadze, G.I. Heat capacity changes upon burial of polar and non polar groups in proteins. Protein Sci. 2001, 10, 1343–1352. [Google Scholar]
- Kumar, S.; Tsai, C.J.; Nussinov, R. Maximal stabilities of reversible two-state proteins. Biochemistry. 2002, 41, 5359–5374. [Google Scholar] [CrossRef]
- Feller, G. Protein stability and enzyme activity at extreme biological temperatures. J. Phys. Condens. Matter 2010, 22, 32–49. [Google Scholar] [CrossRef]
- Chakravarty, S.; Varadarajan, R. Elucidation of factors responsible for enhanced thermal stabilitiy of protein: A structural genomics based study. Biochemistry 2002, 41, 8152–8161. [Google Scholar] [CrossRef]
- D’Amico, S.; Gerday, C.; Feller, G. Dual effects of an extra disulfide bond on the activity and stability of a cold-adapted α-amylase. J. Biol. Chem. 2002, 277, 46110–46115. [Google Scholar] [CrossRef]
- Cipolla, A.; D’Amico, S.; Barumandzadeh, R.; Matagne, A.; Feller, G. Stepwise adaptations to low temperature as revealed by multiple muatnts of psychrophilic α-amylase from Antarctic bacterium. J. Biol. Chem. 2011, 286, 38348–38555. [Google Scholar]
- Papaleo, E.; Pasi, M.; Tiberti, M.; De Gioia, L. Molecular dynamics of mesophilic-like mutants of a cold-adapted enzyme: Insights into distal effects induced by the mutations. PLoS One 2011, 6, e24214. [Google Scholar]
- Miyazaki, K.; Wintrode, P.L.; Grayling, R.A.; Rubingh, D.N.; Arnold, F.H. Directed evolution study of temperature adaptation in a psychrophilic enzyme. J. Mol. Biol. 2000, 297, 1015–1026. [Google Scholar] [CrossRef]
- Taguchi, S.; Komada, S.; Momose, H. The complete amino acid substitutions at position 131 that are positively involved in cold adaptation of subtilisin BPN’. Appl. Environ. Microbiol. 2000, 66, 1410–1415. [Google Scholar] [CrossRef]
- Zong, C.Q.; Song, S.; Fang, N.; Liang, X.; Zhu, H.; Tang, X.; Tang, B. Improvement of low-temperature caseinolytic activity of a thermophilic subtilase by directed evolution and site-directed mutagenesis. Biotechnol. Bioeng. 2009, 104, 862–870. [Google Scholar] [CrossRef]
- Bae, E.; Phillips, G.N. Structure and analysis of highly homologous psychrophilic, mesophilic and thermophilic adenylate kinases. J. Biol. Chem. 2004, 279, 28202–28208. [Google Scholar] [CrossRef]
- Davlieva, M.; Shammo, Y. Structure and biochemical characterization of an adenylate kinase originating from the psychrophilic organism Marinibacillus. marinus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 751–756. [Google Scholar] [CrossRef]
- Aghajari, N.; Van Petegem, F.; Villeret, V.; Chessa, J.P.; Gerday, C.; Haser, R.; Van Beeumen, J. Crystal structures of a psychrophilic metalloprotease reveal new insights into catalysis by cold-adapted proteases. Proteins 2003, 50, 636–647. [Google Scholar] [CrossRef]
- De Baker, M.; McSweeney, S.; Rasmussen, H.B.; Riise, B.W.; Lindley, P.; Hough, E. 9 Å crystal structure of heat-labile shrimp alkaline phosphatase. J. Mol. Biol. 2002, 318, 1265–1274. [Google Scholar] [CrossRef]
- Wang, E.; Koutsioulis, D.; Leiros, H.K.; Andersen, O.A.; Bouriotis, V.; Hough, E.; Heikinheimo, P. Crystal structure of alkaline phosphatase from the Antarctic bacterium TAB5. J. Mol. Biol. 2007, 366, 1318–1331. [Google Scholar] [CrossRef]
- Helland, R.; Leiros, I.; Berglund, G.I.; Willassen, N.P.; Smalas, A.O. The crystral structure of anionic salmon trypsin in complex with bovine pancreatic trypsin inhibitor. Eur. J. Biochem. 1998, 256, 317–324. [Google Scholar]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structures of the psychrophilic Alteromonas. haloplanctis alpha-amylase give insights into cold adaptation at a molecular level. Structure 1998, 6, 1503–1516. [Google Scholar] [CrossRef]
- De Vos, D.; Xu, Y.; Hulpiau, P.; Vergauwen, B.; Van Beeumen, J.J. Structural investigation of cold activity and regulation of aspartate carbamoyltransferase from the extreme psychrophilic bacterium Moritella. profunda. J. Mol. Biol. 2007, 365, 379–395. [Google Scholar] [CrossRef]
- Skalova, T.; Dohnalek, J.; Spiwok, V.; Lipovova, P.; Vondrackova, E.; Petrokova, H.; Duskova, J.; Strnad, H.; Kralova, B.; Hasek, J. Cold-active beta-galactosidase from Arthrobacter. sp. C2–2 forms compact 660 kDa hexamers: Crystal structure at 1.9 Å resolution. J. Mol. Biol. 2005, 353, 282–294. [Google Scholar] [CrossRef]
- Riise, E.K.; Lorentzen, M.S.; Helland, R.; Smalas, A.O.; Leiros, H.-K.S.; Willassen, N.P. The first structure of a cold-active catalase from Vibrio. salmo nicida at 1.96 Å reveals structural aspects of cold adaptation. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 135–148. [Google Scholar]
- Violot, S.; Aghajari, N.; Czjzek, M.; Feller, G.; Sonan, G.; Gouet, P.; Gerday, C.; Haser, R.; Receveur-Bréchot, V. Structure of a full length psychrophilic cellulase from Pseudoalteromonas. haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. J. Mol. Biol. 2005, 348, 1211–1224. [Google Scholar] [CrossRef]
- Russell, R.J.; Gerike, U.; Danson, M.J.; Hough, D.W.; Taylor, G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure 1998, 6, 351–361. [Google Scholar] [CrossRef]
- Berglund, G.I.; Willassen, N.P.; Hordvik, A.; Smalas, A.O. Structure of native pancreatic elastase from North Atlantic salmon at 1.61 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 1995, 51, 925–937. [Google Scholar] [CrossRef]
- Brzuszkiewicz, A.; Nowak, E.; Dauter, Z.; Dauter, M.; Cieslinski, H.; Dlugolecka, A.; Kur, J. Structure of EstA esterase from psychrotrophic Pseudoalteromonas. sp. 643A covanently inhibited by monoethylphosphonate. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 862–865. [Google Scholar] [CrossRef]
- Uppenberg, J.; Hansen, M.T.; Paktar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef]
- Kim, S.Y.; Hwang, K.Y.; Kim, S.H.; Sung, H.C.; Han, Y.S.; Cho, Y. Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium. arcticum. J. Biol. Chem. 1999, 274, 11761–11767. [Google Scholar]
- Karlsen, S.; Hough, E.; Olsen, R.L. Structure and proposed amino-acid sequence of a trypsin from Atlantic cod (Gadus. morhua). Acta Crystallogr. D Biol. Crystallogr. 1998, 54, 32–46. [Google Scholar] [CrossRef]
- Leiros, H.K.; Pey, A.L.; Innselset, M.; Moe, E.; Leiros, I.; Steen, I.H.; Martinez, A. Structure of phenylalanine hydroxylase from Colwellia. psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J. Biol. Chem. 1997, 282, 21973–21986. [Google Scholar]
- Helland, R.; Larsen, A.N.; Smalas, A.O.; Willassen, N.P. Å crystal structure of a proteinase K-like enzyme from a psychrotroph Serratia. species. FEBS. J. 2006, 273, 61–71. [Google Scholar] [CrossRef]
- Tsuruta, H.; Mikami, B.; Aizono, Y. Crystal structure of cold-active protein-tyrosine phosphatase from a psychrophile, Shewanella. sp. J. Biochem. 2005, 137, 69–77. [Google Scholar] [CrossRef]
- Alterio, V.; Aurilia, V.; Romanelli, A.; Parracino, A.; Saviano, M.; D’Auria, S.; De Simone, G. Crystal structure of an S-formyl glutathione hydrolase from Pseudoalteromonas. haloplanktis TAC 125. Biopolymers 2010, 93, 669–677. [Google Scholar]
- Arnorsdottir, J.; Kristjansson, M.M.; Ficner, R. Crystal structure of a subtilisin-like serine proteinase from a psychrotrophic Vibrio. species reveals structural aspects of cold adaptation. FEBS. J. 2005, 272, 832–845. [Google Scholar] [CrossRef]
- Almog, O.; Gonzalez, A.; Godin, N.; de Leeuw, M.; Mekel, M.J.; Klein, D.; Braun, S.; Shoham, G.; Walter, R.L. The crystal structure of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state. Proteins 2009, 74, 489–496. [Google Scholar] [CrossRef]
- Pedersen, H.L.; Willassen, N.P.; Leiros, I. The first structure of a cold-adapted superoxide dismutase (SOD): Biochemical and structural characterization of iron SOD from Aliivibrio. samonicida. Acta Crystallogr. Sect. F Biol. Cryst. Commun. 2009, 65, 84–92. [Google Scholar] [CrossRef]
- Toyota, E.; Ng, K.K.; Kuninaga, S.; Sekizaki, H.; Itoh, K.; Tanizawa, K.; James, M.N. Crystal structure and nucleotide sequence of an anionic trypsin from chum salmon (Oncorrhynchus. keta) in comparison with Atlantic salmon (Salmo. salar) and bovine trypsin. J. Mol. Biol. 2002, 324, 391–397. [Google Scholar] [CrossRef]
- Leiros, I.; Moe, E.; Lanes, O.; Smalas, A.O.; Willassen, N.P. The structure of uracil-DNA glycosylase from Atlantic cod (Gadus. morhua) reveals cold-adaptation features. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 1357–1365. [Google Scholar] [CrossRef]
- Van Petegem, F.; Collins, T.; Meuwis, M.A.; Gerday, C.; Feller, G.; Van Beeumen, J. The structure of a cold-adapted family 8 xylanase at 1.3 Å resolution. Structural adaptations to cold and investigation of the active site. J. Biol. Chem. 2003, 278, 7531–7539. [Google Scholar]
- Hoffman, A.; Bukau, B.; Kramer, G. Structure and function of the molecular chaperone, trigger factor. Biophys. Biochim. Acta 2010, 1803, 650–661. [Google Scholar]
- Tartaglia, G.G.; Dobson, C.M.; Hartl, F.U. Physicochemical determinants of chaperone requirements. J. Mol. Biol. 2010, 400, 579–588. [Google Scholar] [CrossRef]
- Nölting, B.; Salimi, N.; Guth, U. Protein folding forces. J. Theor. Biol. 2008, 251, 331–347. [Google Scholar] [CrossRef]
- Gianni, S.; Ivarsson, Y.; Jemth, P.; Brunori, M.; Travaglini-Allocatelli, C. Identification and characterization of protein folding intermediates. Biophys. Chem. 2007, 128, 105–113. [Google Scholar] [CrossRef]
- Xie, B.; Bian, F.; Chen, X.; He, H.; Guo, J.; Gao, X.; Zeng, Y.; Chen, B.; Zhou, B.; Zhang, Y. Cold adaptation of zinc metalloprotease in the thermolysin family from deep sea and Arctic sea ice bacteria revealed by catalytic and structural properties and molecular dynamics. J. Biol. Chem. 2009, 284, 9257–9269. [Google Scholar]
- Mereghetti, P.; Riccardi, L.; Brandsdal, B.O.; Fantucci, P.; De Gioia, L.; Papaleo, E. Near native-state conformational landscape of psychrophilic and mesophilic enzymes: Probing the folding funnel model. J. Phys. Chem. B. 2010, 114, 7609–7619. [Google Scholar]
- Ferrer, M.; Chernikova, T.N.; Yakimov, M.M.; Golyshin, P.N.; Timmis, K.N. Chaperonins govern growth of Escherichia coli at low temperatures. Nat. Biotechnol. 2003, 21, 1266–1267. [Google Scholar] [CrossRef]
- Ferrer, M.; Lunsdorf, H.; Chernikova, T.N.; Yakimov, M.; Timmis, K.N.; Golyshin, P.N. Functional consequences of single: Double ring transitions in chaperonins: Life in the cold. Mol. Microbiol. 2004, 53, 167–182. [Google Scholar] [CrossRef]
- Goodchild, A.; Saunders, N.F.; Erlan, H.; Raftery, M.; Guilhaus, M.; Curmi, P.M.; Cavicchioli, R. A Proteomic determination of cold-adaptation in the Antarctic archaeon, Methanococcoides. burtonii. Mol. Microbiol. 2004, 53, 309–321. [Google Scholar] [CrossRef]
- Piette, F.; D’Amico, S.; Struvay, C.; Mazzuchelli, G.; Renaut, J.; Tutino, M.L.; Danchin, A.; Leprince, P.; Feller, G. Proteomics of life of low temperatures: Trigger factor is the primary chaperone in the Antarctic bacterium Pseudoalteromonas. haloplanktis TAC 125. Mol. Microbiol. 2010, 76, 120–132. [Google Scholar] [CrossRef]
- Ting, L.; Williams, T.J.; Cowley, M.J.; Lauro, F.M.; Guilhaus, M.; Raftery, M.J.; Cavicchioli, R. Cold adaptation in the marine bacterium Sphingopyxis. alaskensis assessed using quantitative proteomics. Environ. Microbiol. 2010, 12, 2658–2676. [Google Scholar]
- Mykytczuk, N.C.; Trevors, J.T.; Foote, S.J.; Leduc, L.G.; Ferroni, G.D.; Twine, S.M. Proteomics insights into cold adaptation of psychrotrophic and mesophilic Acidothiobacillus. ferrooxidans strains. Van Leeuwenhoek 2011, 100, 259–277. [Google Scholar] [CrossRef]
- Kandror, O.; Goldberg, A.L. Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc. Natl. Acad. Sci. USA 1997, 94, 4978–4981. [Google Scholar] [CrossRef]
- Kramer, G.; Patzelt, H.; Rauch, T.; Kurtz, T.A.; Vorderwulbecke, S.; Bukau, B.; Deurling, E. Trigger factor peptidyl-prolyl cis/trans isomerase activity is not essential for the folding of cytosolic proteins in Escherichia coli. J. Biol. Chem. 2004, 14, 14165–14170. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gerday, C. Psychrophily and Catalysis. Biology 2013, 2, 719-741. https://doi.org/10.3390/biology2020719
Gerday C. Psychrophily and Catalysis. Biology. 2013; 2(2):719-741. https://doi.org/10.3390/biology2020719
Chicago/Turabian StyleGerday, Charles. 2013. "Psychrophily and Catalysis" Biology 2, no. 2: 719-741. https://doi.org/10.3390/biology2020719




