A Brief History of Research on Mitotic Mechanisms
Abstract
:1. Discoveries about Mitosis from Early Descriptions of Mitotic Structures
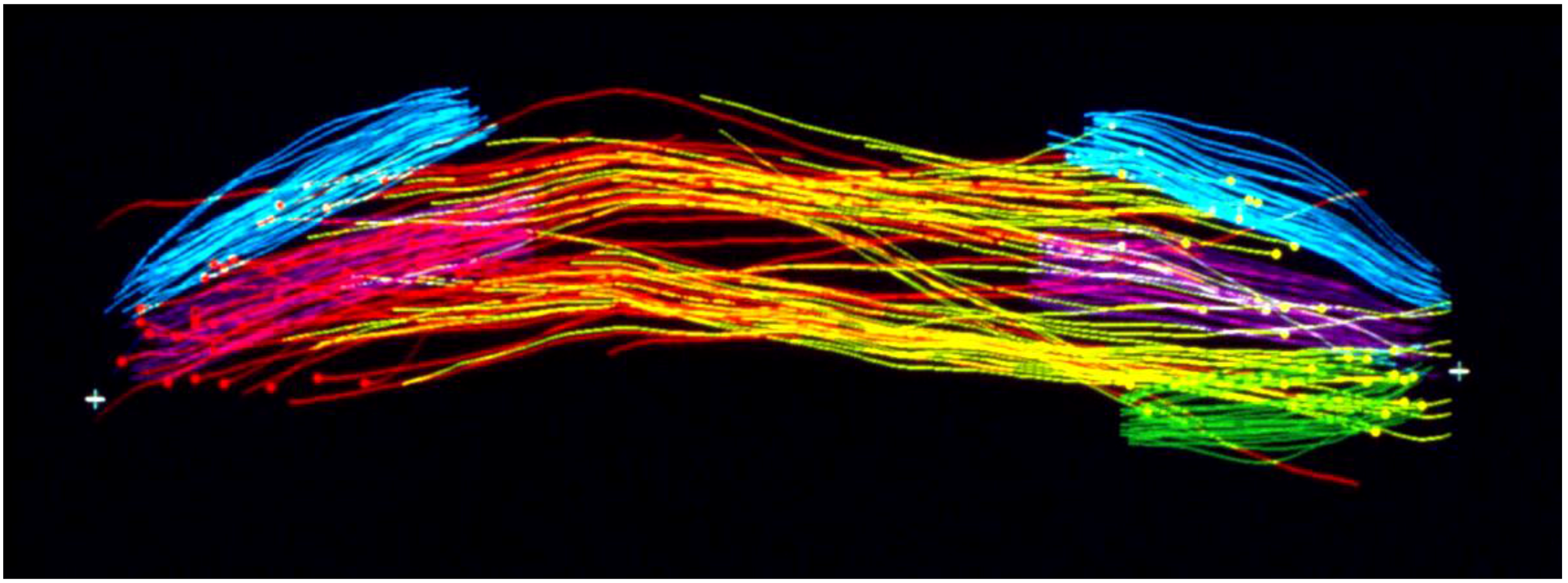
2. New Technologies for Structural Studies Advanced Our Understanding of Spindle Organization
3. Comparisons of Spindles across Phyla
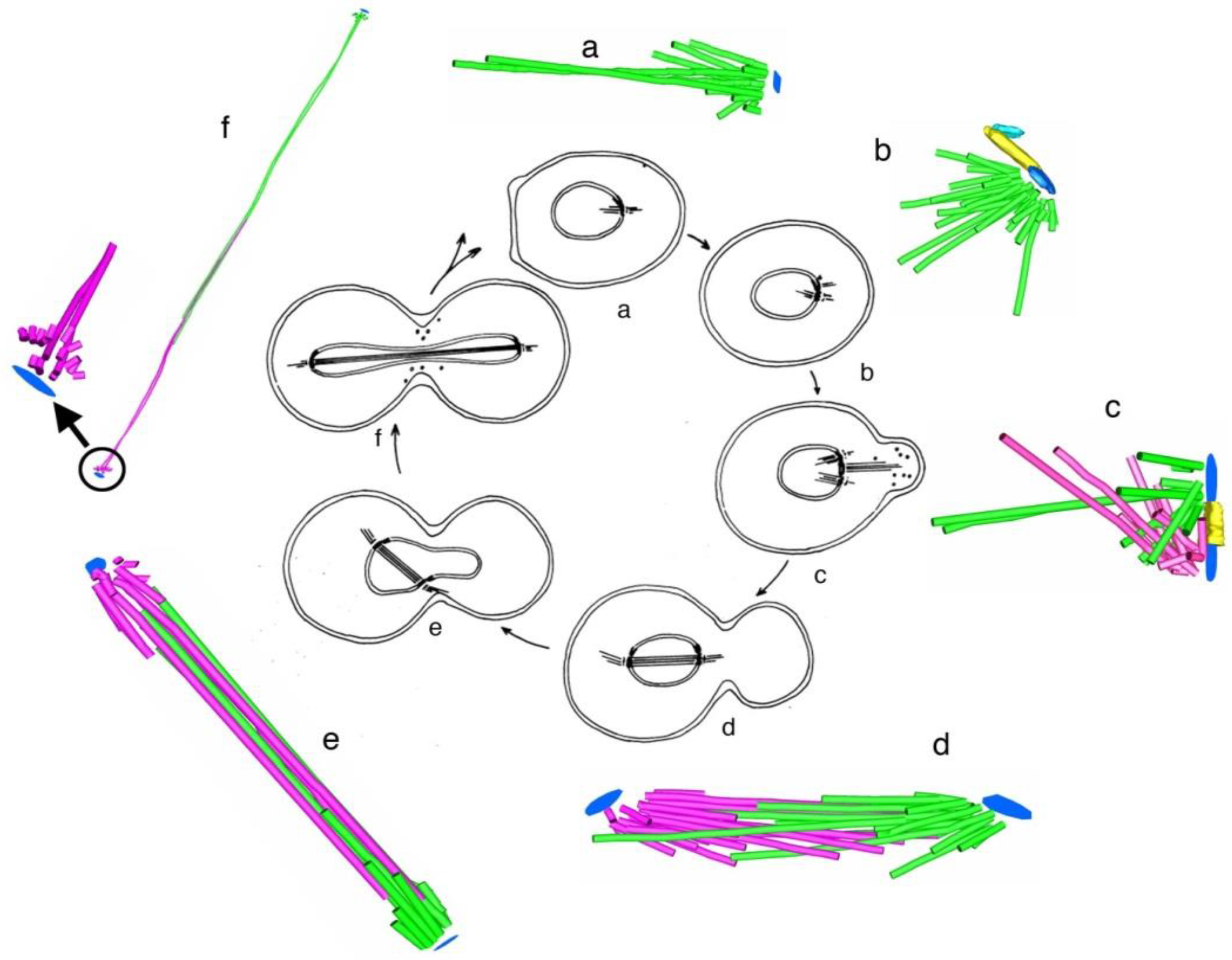
4. A Summary Description of Mitotic and Meiotic Events
5. Biochemical Work to Characterize the Mitotic Machinery
6. Spindle Genetics as a Route to Understanding Mitotic Mechanism
7. Insights into Mitotic Mechanism from Studies of Mitotic Physiology In Vivo
- a.
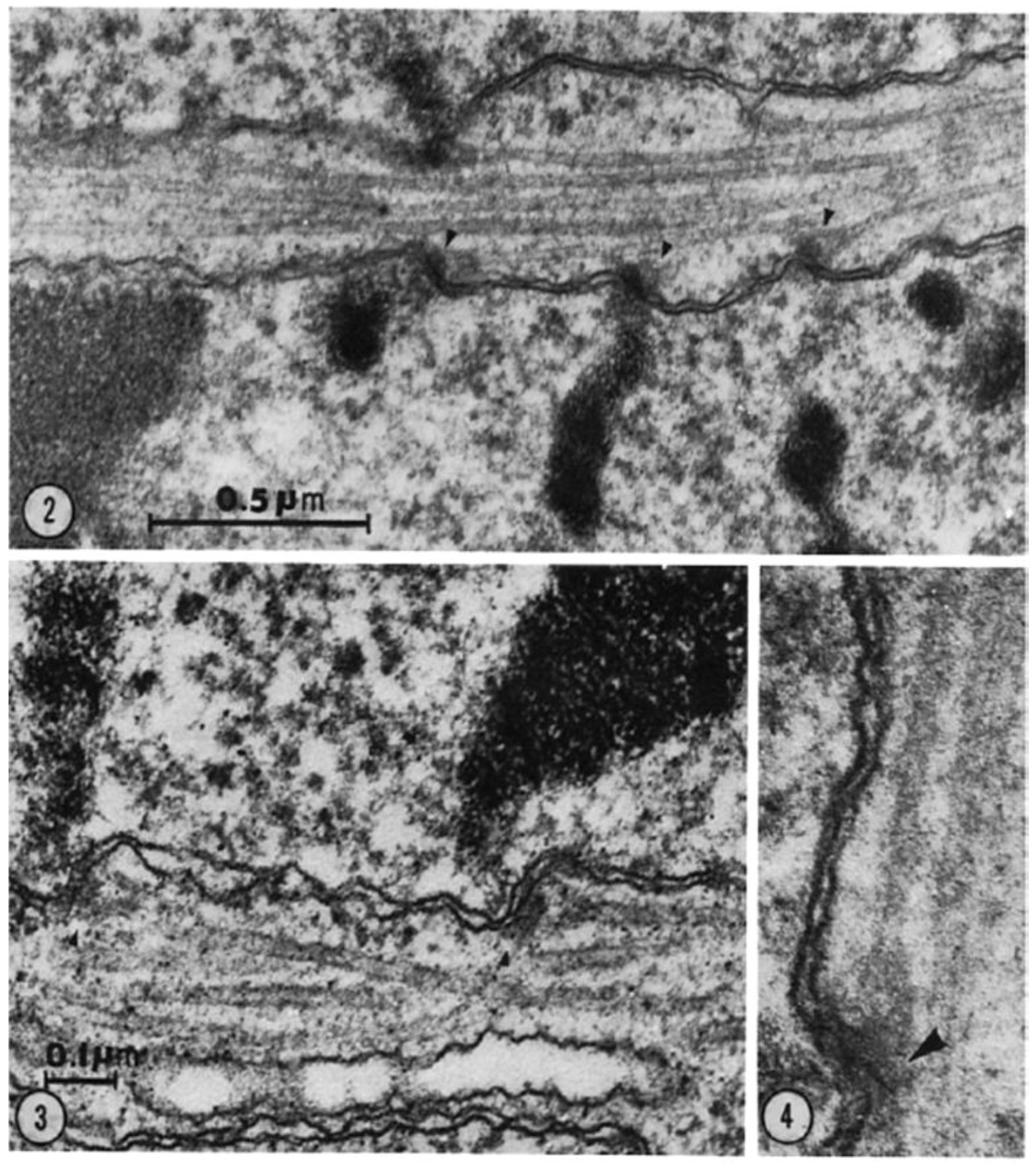
- Experiments on Kinetochores. Improved microscopy enabled descriptions of both localized and diffuse kinetochores, as seen in certain insects [121]. Experimental work using X-rays to fragment chromosome [122,123] provided direct evidence for the role of kinetochores in spindle attachment and chromosome segregation. X-ray induced fragmentation of chromosomes with localized kinetochore produced multiple fragments, only one of which contained a kinetochore. This fragments attached to the spindle and moved normally, but the fragments that lacked a kinetochore failed to attach and were lost at subsequent divisions [124]. By contrast, chromosomes with diffuse kinetochores were connected to spindle fibers by the entire poleward surface of each chromosome. X-ray-induced fragments of these chromosomes retained kinetic capacity; regardless of how small the pieces became, each chromosome fragment was pulled to opposite spindle poles by its associated kinetochore fibers [123]. This work established the importance of kinetochores in chromosome motion.
- b.
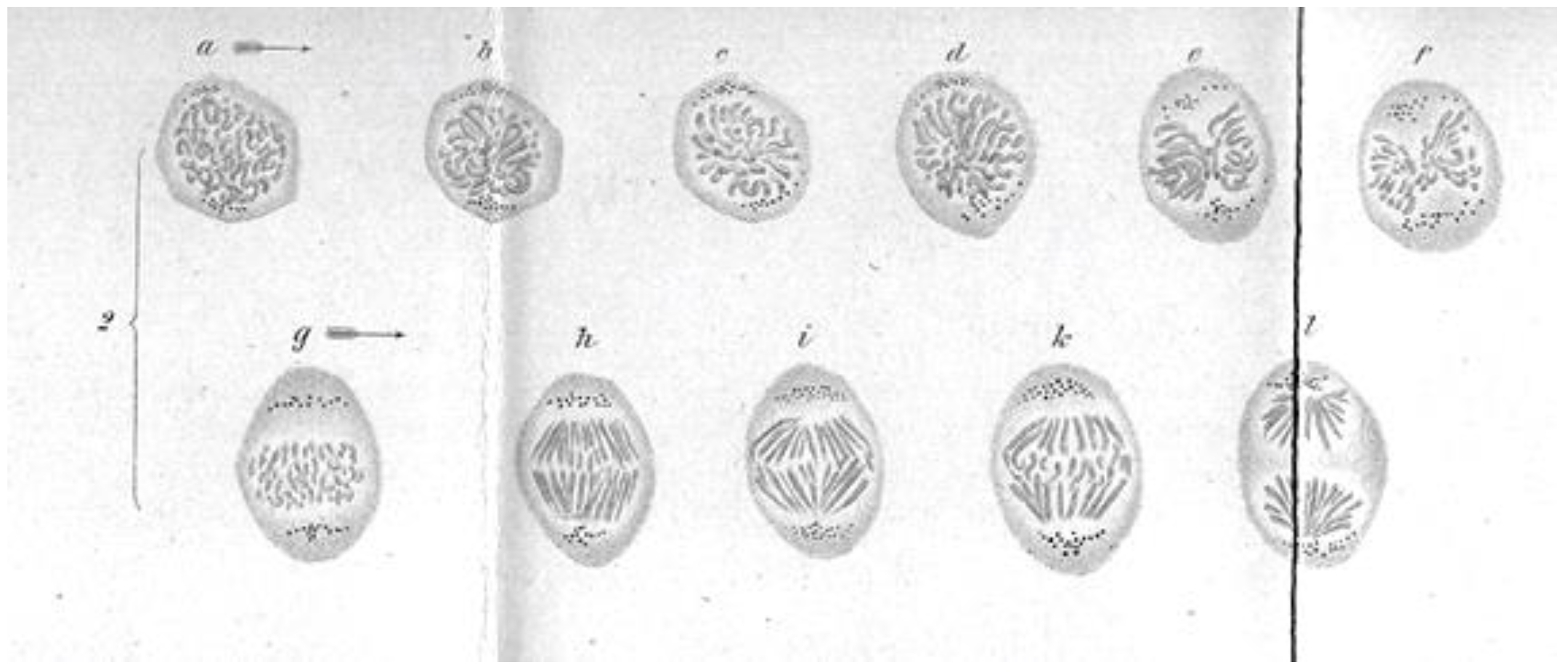
- Observations and Experiments on Chromosome Movements. Some diversity in anaphase was discovered by quantitative descriptions of chromosome movements in living cells. Changes in spindle length and kinetochore separation revealed two phases of chromosome movement [25,125]. One involved the shortening of chromosomal fibers, the other an elongation of the spindle with consequent chromosome movement. Early descriptive work revealed variation in the extent to which organisms relied on Anaphase A or B. Some used only Anaphase A (Tradescantia) and some only Anaphase B (Primary spermatocytes of the Aphid, Tamalia [125]). Some cells used both though separated in time (Secondary spermatocytes and embryonic cells of the Aphid, Tamalia; Hemiptera and Homoptera; [125]), and others used both anaphase mechanisms overlapping in time and therefore difficult to distinguish (grasshoppers; chick tissue culture cells [25]).
- c.
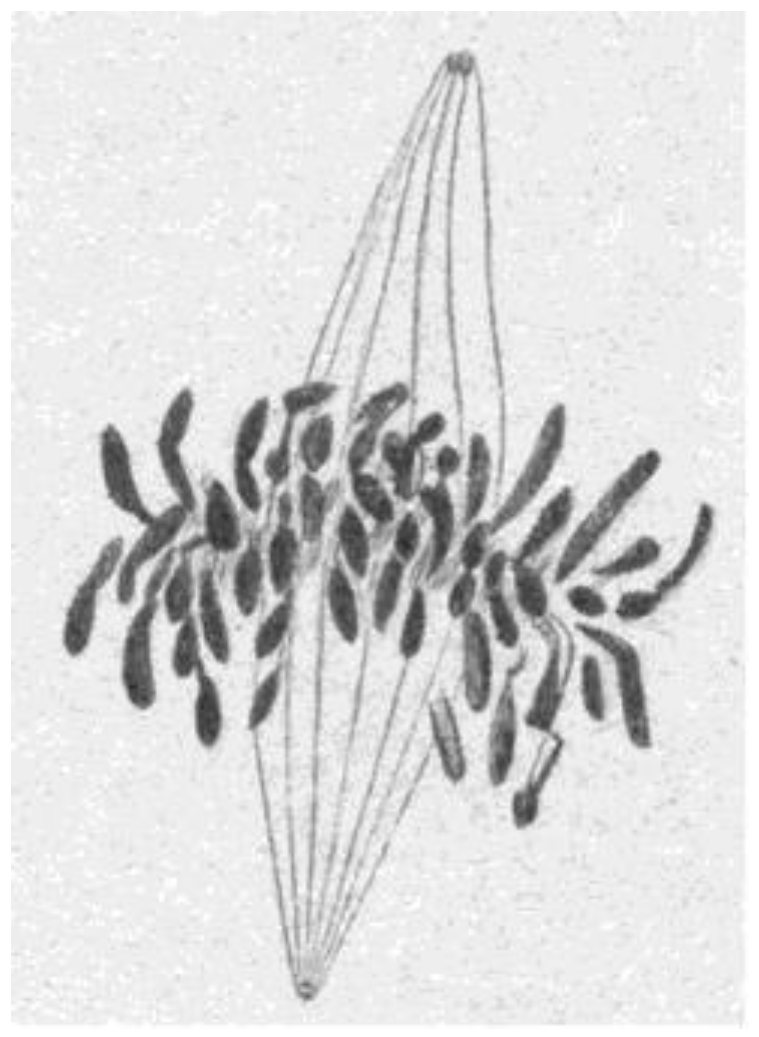
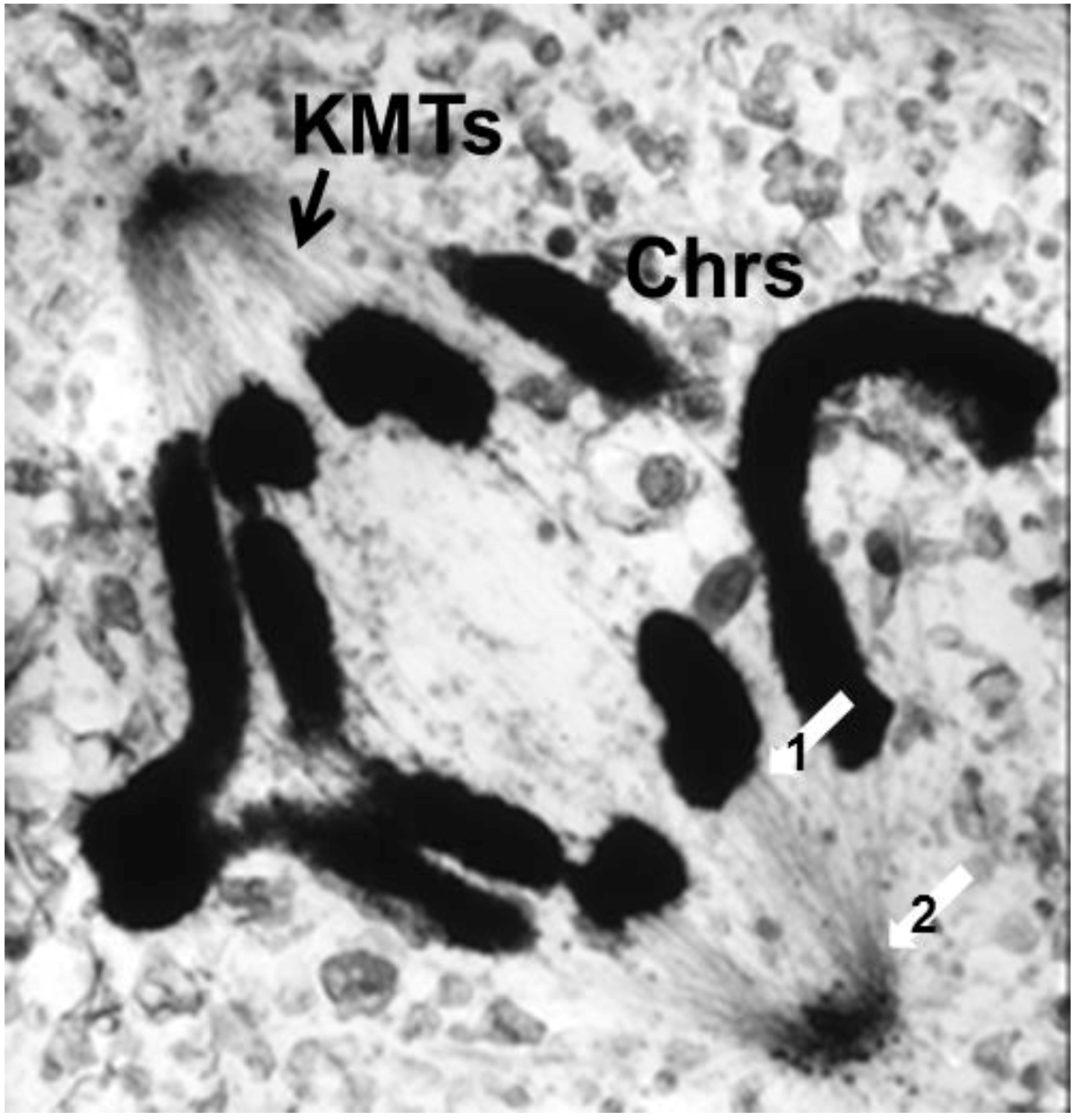
- Chromosome Pulling Forces Were Detected in Prometaphase. Descriptions of chromosome behavior in a variety of meiotic cells revealed a “pre-metaphase stretch” in which the distance between kinetochores on homologous chromosomes was greatly extended by action of the spindle [131,132], e.g., mantids and phasmids (Figure 18). This pre-metaphase stretch was followed by the gradual re-contraction of chromosomes and a resumption of normal prometaphase congression. Notably, the timing of the stretch and the resumption of congression were asynchronous on adjacent chromosomes, implying that mitotic forces acted independently on individual chromosomes. Taken together, these studies suggested the autonomy of chromosome movements during mitosis, eliminating models of collective transport by the spindle. These observations contributed to the rise of traction fiber models, in which kinetochores and their associated chromosomes were pulled individually towards the spindle poles to which they were attached.
- d.
- Physical Perturbations of the Spindle as a Whole. Some of the most convincing evidence for the existence of chromosomal fibers in vivo came from experimental manipulations of dividing cells. The centrifugation of mitotic cells distorted the spindle [121], stretching it [133] and/or severing chromosome attachment sites [134]. These perturbations also separated anaphase spindles into two half spindles, showing that chromosome-spindle pole attachments were strong enough to resist the centrifugal forces that distorted bivalent chromosomes. Centrifugation experiments further suggested a gel-like mechanical texture of the spindle.
- e.
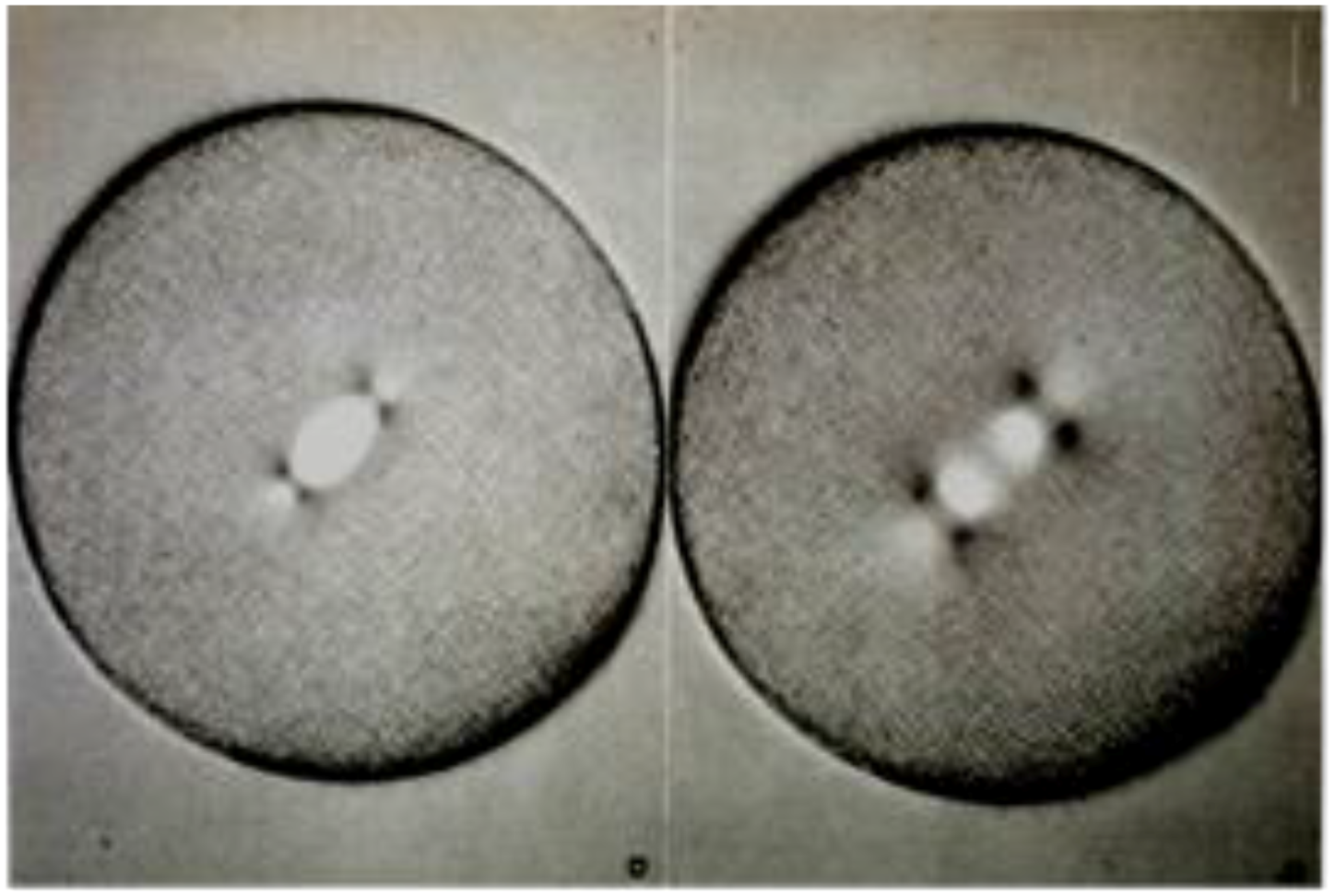
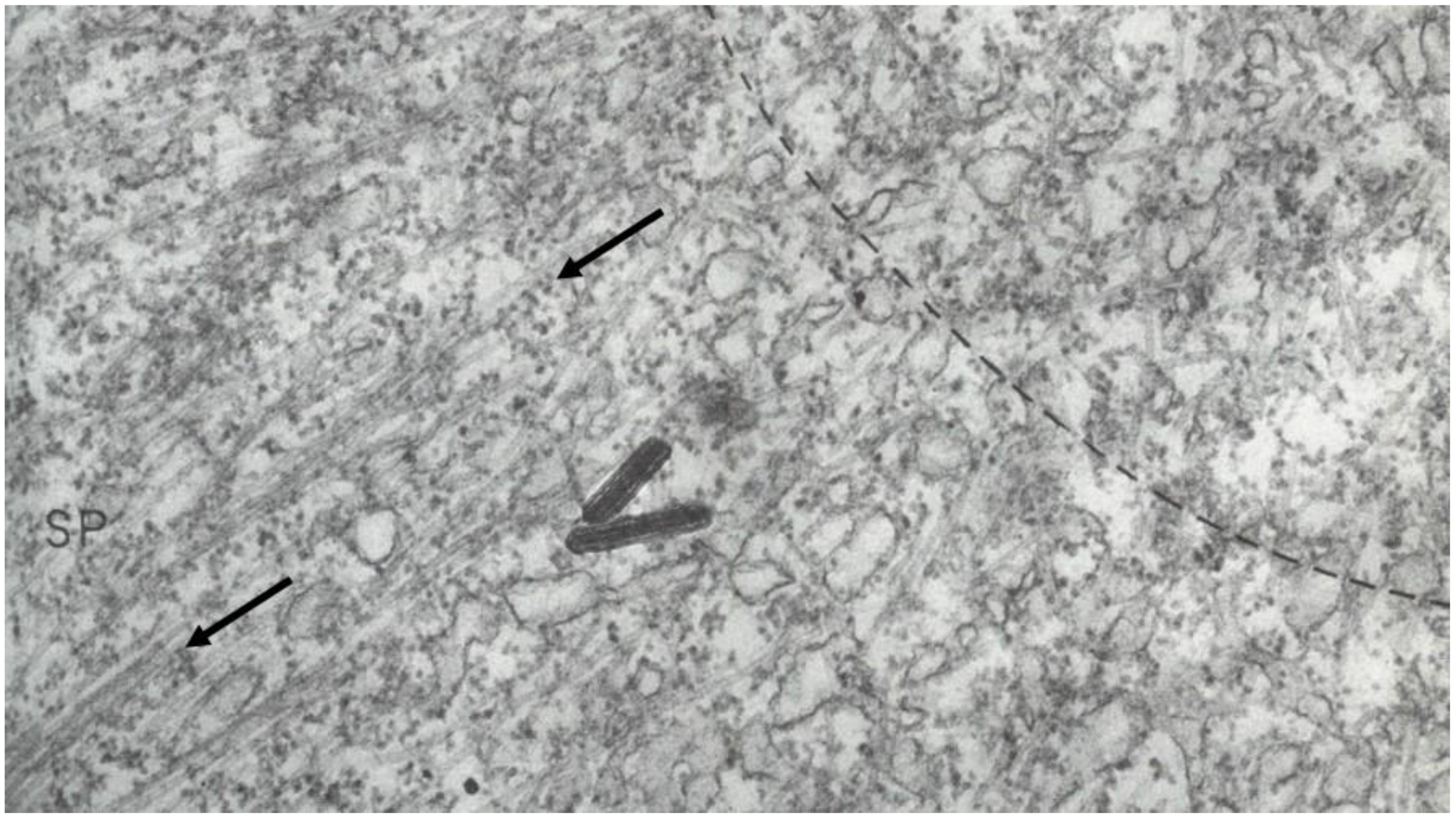
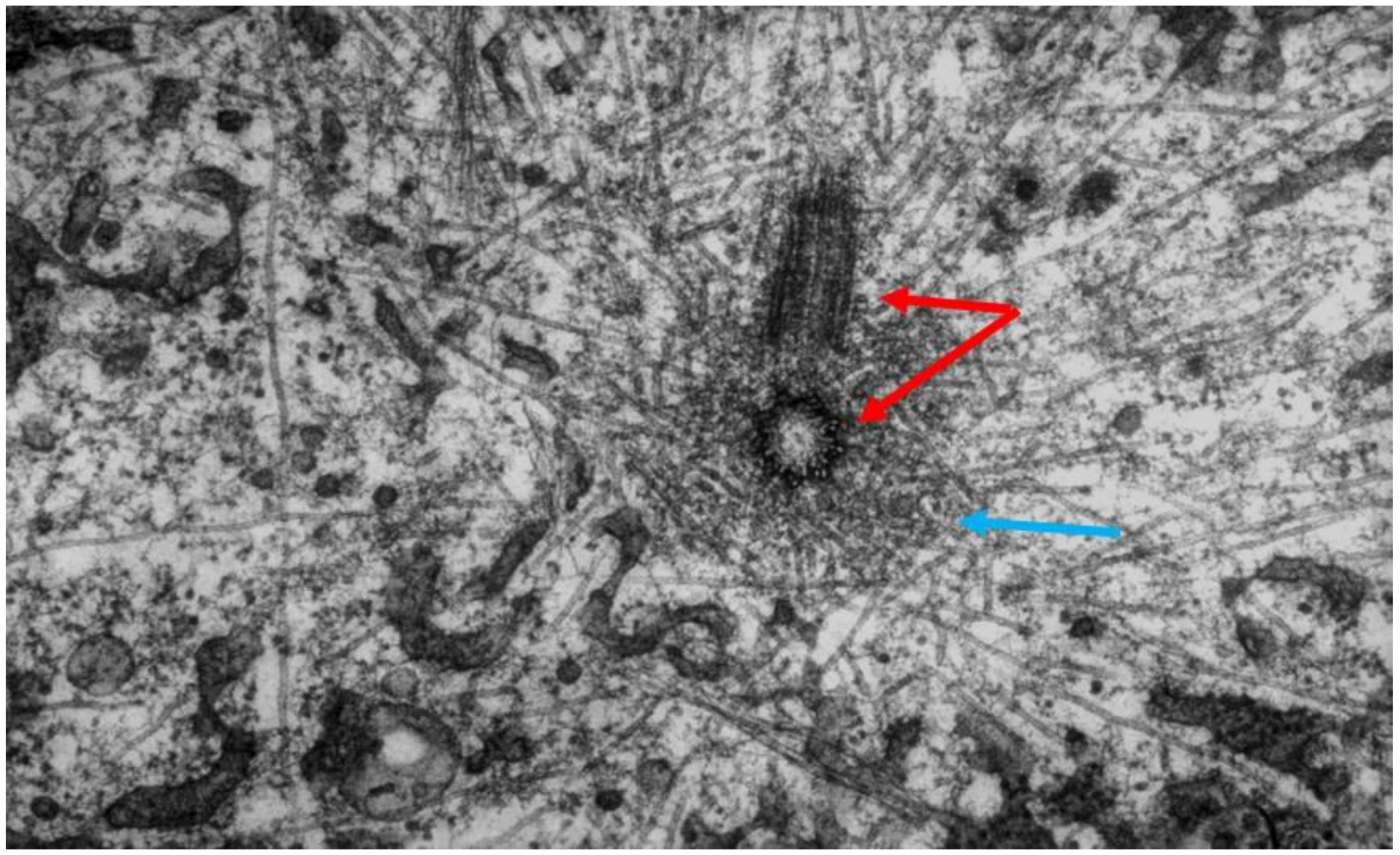
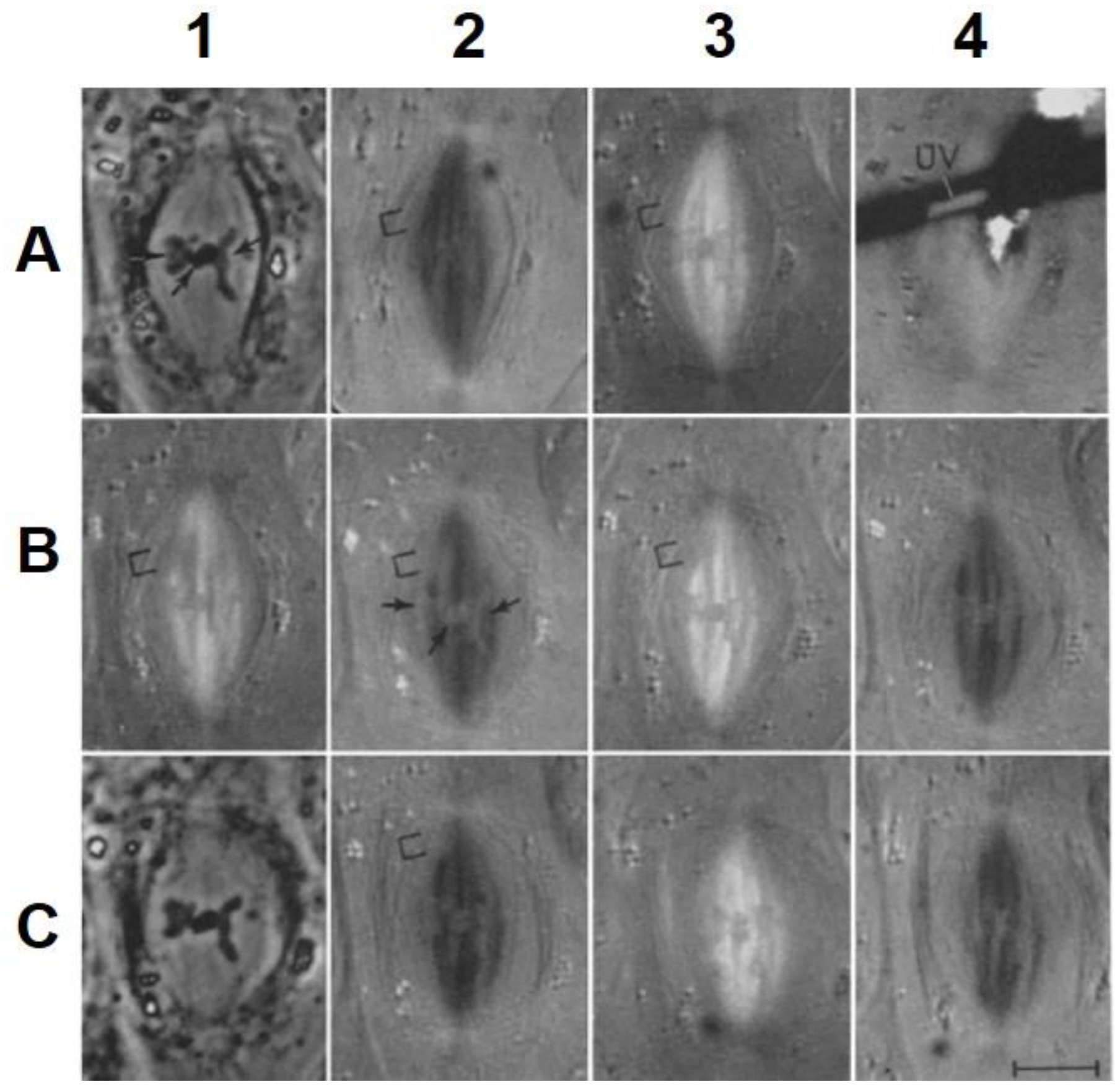
- Local Perturbations of Spindle Structure and Function. Experiments with microbeams of ultraviolet (UV) light raised provocative questions about the nature of MT dynamics and the roles of MTs in chromosome movements. Forer [144] reported that UV microbeam irradiation of chromosomal fibers in crane fly spermatocytes at metaphase produced localized “areas of reduced birefringence” (ARBs). ARBs subsequently moved poleward, even while the chromosomes remained aligned at the metaphase plate. Such observations indicated that chromosomal fibers were not static and suggested a continuous poleward flux of materials within the chromosomal fiber (Figure 19). Moreover, in anaphase, Forer reported variability in the impact of ARBs on chromosome movement [144]. Two thirds of the time, chromosome to pole movements ceased when irradiated fibers contained ARBs. One third of the ARBs, however, had no impact on chromosome movement. Moreover, two thirds of irradiated fibers did not develop ARBs, yet chromosome to pole movements were blocked as frequently as when an ARB formed. Forer interpreted these results to mean that chromosomal fibers contained two components: birefringent MTs, which neither produce nor transmit the forces required for chromosome movements, and a non-birefringent element that was required for traction the forces that pulled chromosomes poleward in anaphase A. Electron microscopy later confirmed that in at least some experiments, MTs within the ARBs were severed and/or depolymerized [145,146]. Forer’s complicated results from anaphase spindles have never really been explained.
- f.
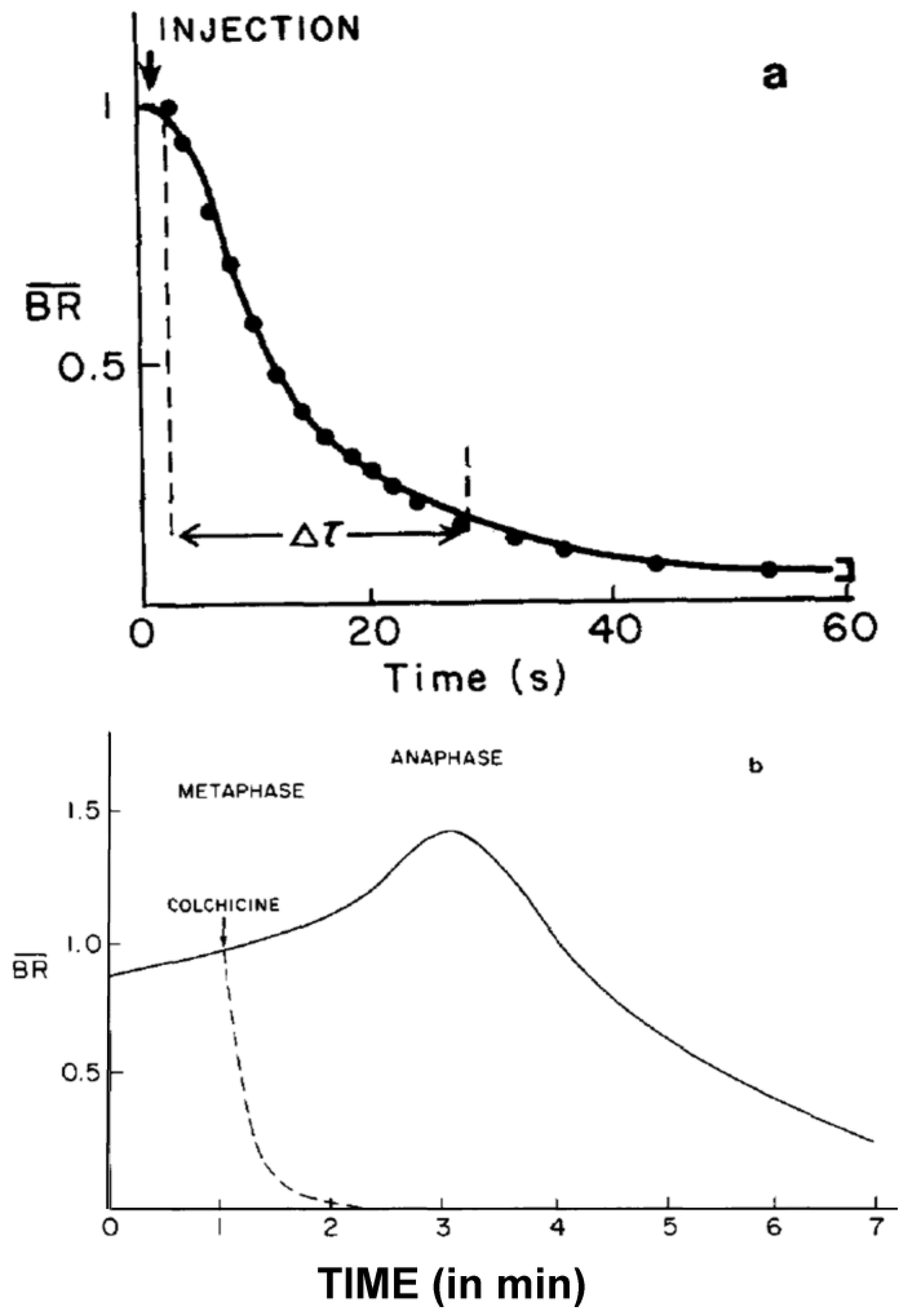
- Studies on MT Dynamics in Vivo. Salmon and coworkers [149] microinjected high concentrations of colchicine into dividing sea urchin eggs, blocking spindle MT polymerization, and allowing the rate of depolymerization to be measured. A rapid decrease in birefringence (BR) reflected the disappearance of non-kinetochore MTs, while kinetochore MTs were again differentially stable (Figure 20). Surprisingly, the calculated rate of depolymerization (180–992 dimers per second) was significantly faster than predicted from the in vitro parameters of tubulin dynamics. It was, however, consistent with a significantly different, “dynamic instability” pathway concurrently proposed for MT behavior [150]. At about the same time, several investigators capitalized on photobleaching, as well as on the incorporation of injected tubulin and a specially labeled tubulin, using fully functional tubulin “analogues” to interrogate the pathways of tubulin assembly and disassembly during mitosis and the mechanistic basis of chromosome movements [151,152,153].
- g.
- Investigating Spindle-Chromosome Interactions by Micro-Manipulation. In a classic series of studies, Nicklas and coworkers used the tips of fine microneedles to tug on chromosomes in grasshopper spermatocytes and characterize their attachment to the meiotic spindle. They showed that chromosome-attached spindle fibers were essentially inextensible, that they were bound to the rest of the spindle near the pole, and that they were readily displaced, either laterally or toward the pole, without losing their chromosome attachment; moreover, each chromosome was quite independent of its neighbors [158]. Each chromosome behaved like a pendulum suspended by a thin wire and free to swing from a pivot point at the spindle pole. The chromosomal fiber was stiff under tension but flexible to compression and bending. Nicklas further demonstrated that during prometaphase and metaphase chromosomes could be detached from their fibers by repeated tugging with the needle. Once released from the needle, they would reattach to the spindle (See Supplementary Movie 1). In contrast, chromosomes in anaphase could not be detached from their fibers, despite great effort. The segregation of chromosomes following these micromanipulations was indistinguishable from that in control, unmanipulated cells.
- h.
- Assessment of Spindle-Generated Forces. The first studies of mitotic force magnitudes considered the relationship between chromosome size and velocity. These studies showed that velocity was independent of size and thus of viscous load over a limited range. Chromosomes varying more than 2-fold in size exhibit the same speeds in both prometaphase and anaphase [130]. Similarly, McNeil and Berns [166] showed in metaphase PtK2 cells that when a single kinetochore is irradiated with a high powered laser, the unirradiated sister kinetochore transports twice the normal amount of chromatin at the same velocity as a normal anaphase chromosome.
- i.
- Experiments to Investigate Chromosome Congression to the Metaphase Plate. Students of mitosis have long recognized the importance of metaphase in establishing a uniform initial condition for subsequent chromosome segregation. Numerous hypotheses were advanced for how chromosomes are brought to the spindle midplane. A simple and important idea emerged from the studies by Rashevsky [171], Hughes-Schrader [131], and Ostergren [172,173]: the poleward force on a chromosome might increase with distance from the pole. Paired chromosomes would then congress to the spindle equator because that position allowed the opposing forces to be balanced. Considerable evidence supports this force-balance theory, including observations on the consequences of upsetting the force balance. If sister chromatids are disconnected, either naturally at anaphase or artificially at metaphase [174], the opposing forces are uncoupled and each chromatid moves poleward. Similarly, a laser micro-beam can be used to destroy one metaphase kinetochore, and the chromosome then moves towards the pole to which the undamaged kinetochore is attached [166,175]. Thus, prometaphase and metaphase chromosomes are clearly being pulled in two directions at once.
8. Models of Mitotic Mechanisms
9. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Schneider, A. Untersuchungen über Plathelminthen. In Bericht der Oberhessischen Gesellschaft für Natur- und Heilkunde; Upper-Hessian Society for Natural and Medical Science: Giessen, Hesse, Germany, 1873; Volume 14. (In German) [Google Scholar]
- Strasburger, E. Zellbildung und Zelltheilung “Cell Formation and Cell Division”; Gustav Fischer: Jena, Germany, 1880. [Google Scholar]
- Van Beneden, E. Recherches sur les Dicyemides. Bull. Acad. R. 1876, 41, 1–111. [Google Scholar]
- Flemming, W. Zur Kenntniss der Zelle und ihre Lebenserscheinungen. Arch. Mikr. Anat. 1878, 16, 302–436. [Google Scholar] [CrossRef]
- Flemming, W. Contributions to the knowledge of the cell and its vital processes. J. Cell Biol. 1965, 25, 3–69. [Google Scholar] [PubMed]
- Flemming, W. Zellsubstanz, Kern und Zelltheilung; Vogel: Leipzig, Germany, 1882. (In German) [Google Scholar]
- Hooke, R. Micrographia: Or Some Physiological Descriptions of Minute Bodies Made by Magnifying Glasses with Observations and Inquiries Thereupon; James Allestry: London, UK, 1667. [Google Scholar]
- Van Leeuwenhoek, A. Arcana Naturae Detacta; Apud Henricum a Krooneveld: Delft, the Netherlands, 1695. [Google Scholar]
- Virchow, R. Die Cellularpathologie in Ihrer Begründung auf Physiologische und Pathologische Gewebelehre; Verlag von August Hirschwald: Berlin, Germany, 1858. [Google Scholar]
- Abbe, E. A New Illuminating Apparatus for the Microscope. Mon. Microsc. J. 1875, 13, 77–82. [Google Scholar] [CrossRef]
- Mayzel, W. Ueber eigenthümiche Vorgänge bei der Theilung der Kerne in Epithelialzellen” (On peculiar events during the division of nuclei of epithelial cells). Zentralbl. Med. Wiss. 1885, 13, 849–852. [Google Scholar]
- Weissmann, A. Die Continuität des Keimplasma’s als Grundlage einer Theorie der Vererbung; Fischer: Jena, Germany, 1885. [Google Scholar]
- Boveri, T.H. Ergebnisse über die Konstitution der Chromatischen Substanz des Zelkerns; Fisher: Jena, Germany, 1904. [Google Scholar]
- Sutton, W.S. The chromosomes in heredity. Biol. Bull. 1903, 4, 231–251. [Google Scholar] [CrossRef]
- Wilson, E.B. The Cell in Development and Heredity; MacMillan, Inc.: New York, NY, USA, 1925. [Google Scholar]
- Fischer, A. Fixirung, Färbung und Bau des Protoplasmas: Kritische Untersuchungen über Technik und Theorie in der Neueren Zellforschung; Fisher: Jena, Germany, 1899. [Google Scholar]
- Hardy, W.B. On Spindles. J. Physiol. 1899, 24, 158–210. [Google Scholar] [CrossRef] [PubMed]
- Lauterborn, R. Untersuchengen ueber Bau, Kernteilung und Bewegung der Diatomeen; Wilhelm Engelmann: Leipzig, Germany, 1896. [Google Scholar]
- Lauterborn, R. Cell Division in Diatoms. Protoplasma 1984, 120, 132–154. [Google Scholar]
- Chambers, R. Microdissection studies. II. The cell aster, a reversible gelation phenomenon. J. Exp. Zool. 1917, 23, 483–504. [Google Scholar] [CrossRef]
- Zernike, F. How I discovered Phase optics. Science 1955, 121, 345–349. [Google Scholar] [CrossRef]
- Cleveland, L.R.; Hall, S.R.; Sanders, E.P. The Wood-Feeding Roach Cryptocercus, Its Protozoa, and the Symbiosis between Protozoa and Roach, 1st ed.; American Academy of Arts and Sciences: Cambridge, MA, USA, 1934; Volume 17, p. 406. [Google Scholar]
- Schmidt, W.J. Doppelbrechung von Karyoplasma, Metaplasma und Zytoplasma; Gebrueder Borntraeger: Berlin, Germany, 1937. (In German) [Google Scholar]
- Schmidt, F.O. The ultrastructure of protoplasmic constituents. Physiol. Rev. 1939, 19, 270–302. [Google Scholar]
- Hughes, A.F.; Swann, M.M. Anaphase Movements in the Living Cell. J. Exp. Biol. 1948, 25, 45–72. [Google Scholar]
- Inoue, S.; Dan, K. Birefringence of the dividing cell. J. Morph. 1951, 89, 423–456. [Google Scholar] [CrossRef]
- Inoue, S.; Hyde, W.L. Studies on depolarization of light at microscope lens surfaces, II. The simultaneous realization of high resolution and high sensitivity with the polarizing microscope. J. Cell Biol. 1957, 3, 831–838. [Google Scholar] [CrossRef]
- Rozsa, G.; Wyckoff, R.W.G. The electron microscopy of dividing cells. Biochim. Biophys. Acta 1950, 6, 334–339. [Google Scholar] [CrossRef]
- Bernhard, W.; De Harven, E. Electron microscopic study of the ultrastructure of centrioles in vertebra. Z. Zellforsch. Mikrosk. Anat. 1956, 45, 378–398. [Google Scholar] [PubMed]
- Harris, P. Some Observations Concerning Metakinesis in Sea Urchin Eggs. J. Cell Biol. 1965, 25, 73–77. [Google Scholar] [CrossRef]
- Roth, L.E.; Daniels, E.W. Electron microscopic studies of mitosis in amebae: II The Giant Ameba Pelomyxa carolinensis. J. Cell Biol. 1962, 57–78. [Google Scholar] [CrossRef]
- Sabatini, D.D.; Bensch, K.; Barrnett, R.J. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J. Cell Biol. 1963, 17, 19–58. [Google Scholar] [CrossRef] [PubMed]
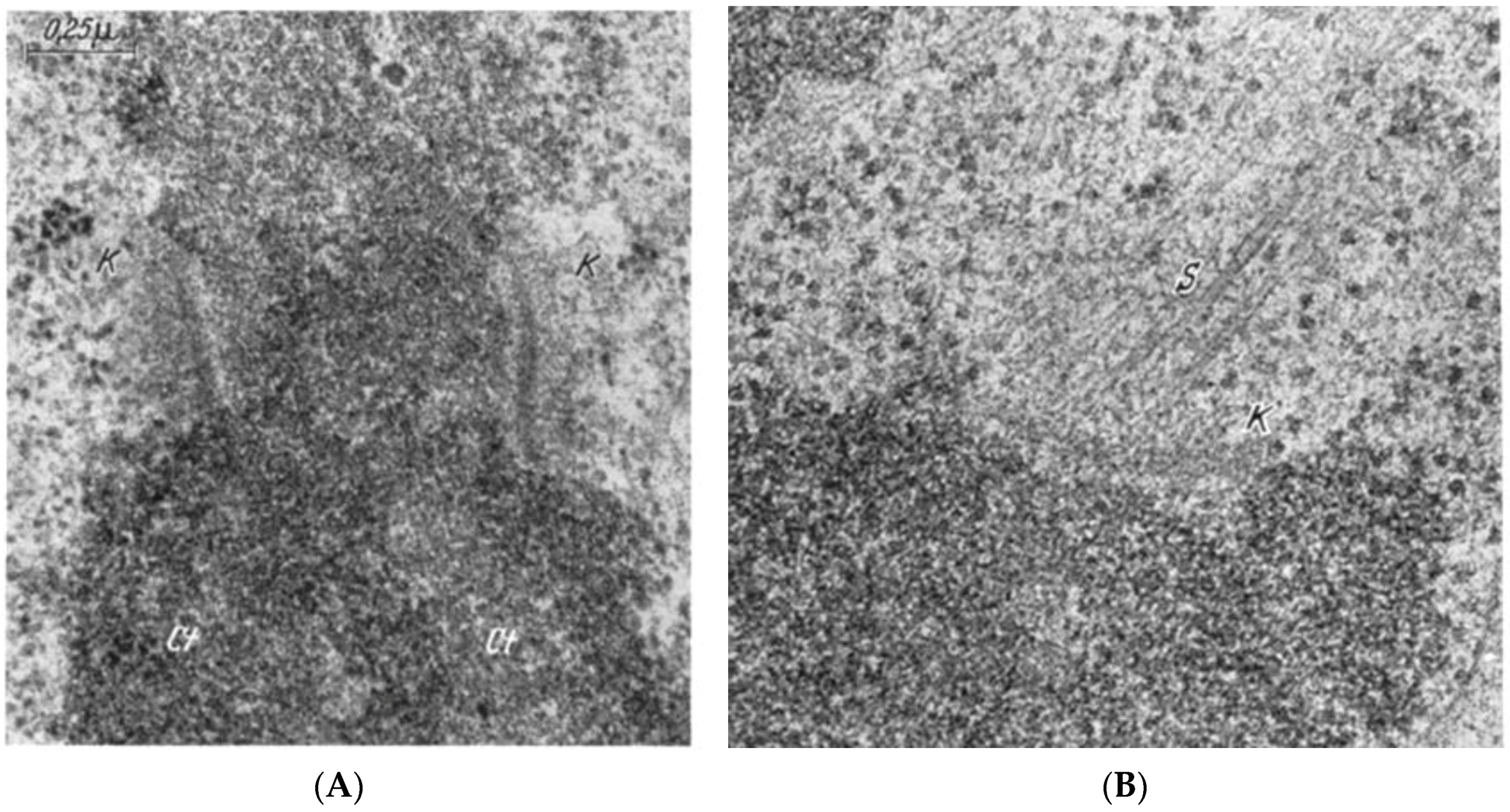
- Brinkley, B.R.; Stubblefield, E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma 1966, 19, 28–43. [Google Scholar] [CrossRef] [PubMed]
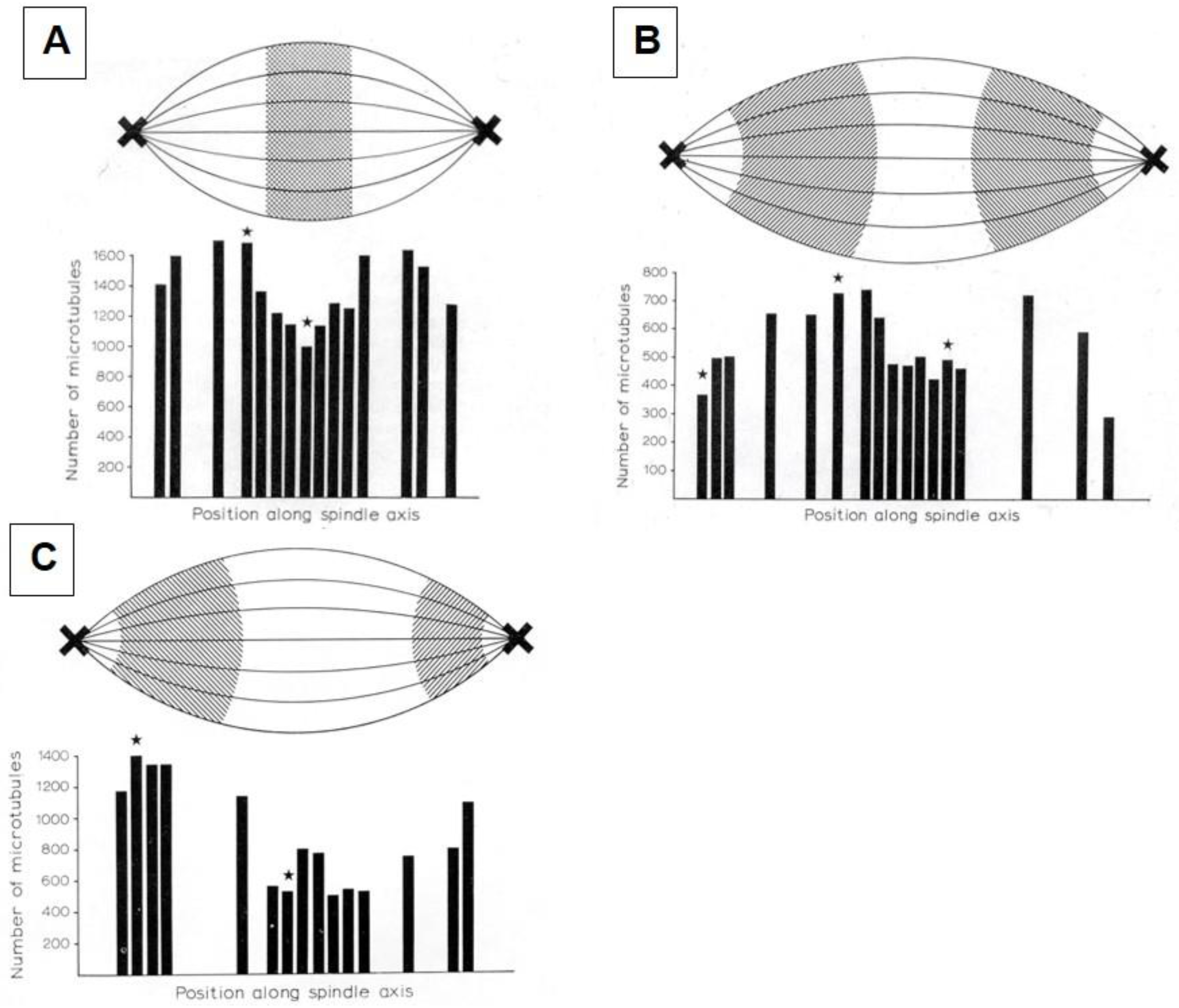
- McIntosh, J.R.; Landis, S.C. The Distribution of Spindle Microtubules during Mitosis in Cultured Human Cells. J. Cell Biol. 1971, 49, 468–497. [Google Scholar] [CrossRef] [PubMed]
- Fuge, H. Microtubule distribution in metaphase and anaphase spindles of the spermatocytes of Pales ferruginea. A quantitative analysis of serial cross-sections (author’s transl). Chromosoma 1973, 43, 109–143. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; Cande, W.Z.; Snyder, J.A.; Vanderslice, K. Studies on the mechanism of mitosis. Ann. N.Y. Acad. Sci. 1975, 253, 383–406. [Google Scholar] [CrossRef]
- McIntosh, J.R.; Cande, W.Z.; Snyder, J.A. Structure and physiology of the mammalian mitotic spindle. Soc. Gen. Physiol. Ser. 1975, 30, 31–76. [Google Scholar] [PubMed]
- Nicklas, R.B.; Kubai, D.F.; Hays, T.S. Spindle microtubules and their mechanical associations after micromanipulation in anaphase. J. Cell Biol. 1982, 95, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.B.; Ris, H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J. Cell Sci. 1976, 22, 219–242. [Google Scholar] [PubMed]
- McDonald, K.; Pickett-Heaps, J.D.; McIntosh, J.R.; Tippit, D.H. On the mechanism of anaphase spindle elongation in Diatoma vulgare. J. Cell Biol. 1977, 74, 377–388. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; Roos, U.P.; Neighbors, B.; McDonald, K.L. Architecture of the microtubule component of mitotic spindles from Dictyostelium discoideum. J. Cell Sci. 1985, 75, 93–129. [Google Scholar] [PubMed]
- Winey, M.; Mamay, C.L.; O’Toole, E.T.; Mastronarde, D.N.; Giddings, T.H.; McDonald, K.L.; McIntosh, J.R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995, 129, 1601–1615. [Google Scholar] [CrossRef] [PubMed]
- Kilmartin, J.V.; Adams, A.E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 1984, 98, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L.; Borisy, G.G. The attachment of kinetochores to the pro-metaphase spindle in PtK1 cells. Recovery from low temperature treatment. Chromosoma 1981, 82, 693–716. [Google Scholar] [CrossRef] [PubMed]
- McDonald, K.L.; O’Toole, E.T.; Mastronarde, D.N.; McIntosh, J.R. Kinetochore microtubules in PTK cells. J. Cell Biol. 1992, 118, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Mastronarde, D.N.; McDonald, K.L.; Ding, R.; McIntosh, J.R. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993, 123, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Amos, L.; Klug, A. Arrangement of subunits in flagellar microtubules. J. Cell Sci. 1974, 14, 523–549. [Google Scholar] [PubMed]
- McIntosh, J.R.; Hepler, P.K.; Van Wie, D.G. Model for Mitosis. Nature 1969, 224, 659–663. [Google Scholar] [CrossRef]
- Pease, D.C. Hydrostatic pressure effects upon the spindle figure and chromosome movements. Biol. Bull. 1946, 91, 145–169. [Google Scholar] [CrossRef] [PubMed]
- Witt, P.L.; Ris, H.; Borisy, G.G. Origin of kinetochore microtubules in Chinese hamster ovary cells. Chromosoma 1980, 81, 483–505. [Google Scholar] [CrossRef] [PubMed]
- Weisenberg, R.; Rosenfeld, A. Role of intermediates in microtubule assembly in vivo and in vitro. Ann. N.Y. Acad. Sci. 1975, 253, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Telzer, B.R.; Moses, M.J.; Rosenbaum, J.L. Assembly of microtubules onto kinetochores of isolated mitotic chromosomes of HeLa cells. Proc. Natl. Acad. Sci. USA 1975, 72, 4023–4027. [Google Scholar] [CrossRef] [PubMed]
- Bergen, L.G.; Borisy, G.G. Head-to-tail polymerization of microtubules in vitro. Electron microscope analysis of seeded assembly. J. Cell Biol. 1980, 84, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Bergen, L.G.; Kuriyama, R.; Borisy, G.G. Polarity of microtubules nucleated by centrosomes and chromosomes of Chinese hamster ovary cells in vitro. J. Cell Biol. 1980, 84, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Heidemann, S.R.; McIntosh, J.R. Visualization of the structural polarity of microtubules. Nature 1980, 286, 517–519. [Google Scholar] [CrossRef] [PubMed]
- Euteneuer, U.; McIntosh, J.R. Polarity of midbody and phragmoplast microtubules. J. Cell Biol. 1980, 87, 509–515. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; Euteneuer, U. Tubulin hooks as probes for microtubule polarity: An analysis of the method and an evaluation of data on microtubule polarity in the mitotic spindle. J. Cell Biol. 1984, 98, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Telzer, B.R.; Haimo, L.T. Decoration of spindle microtubules with Dynein: Evidence for uniform polarity. J. Cell Biol. 1981, 89, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Bajer, A.S.; Mole-Bajer, J. Spindle Dynamics and Chromosome Movement. Int. Rev. Cytol. Suppl. 1972, 34, 1–271. [Google Scholar]
- De Mey, J.; Lambert, A.M.; Bajer, A.S.; Moeremans, M.; De Brabander, M. Visualization of microtubules in interphase and mitotic plant cells of Haemanthus endosperm with the immuno-gold staining method. Proc. Natl. Acad. Sci. USA 1982, 79, 1898–1902. [Google Scholar] [CrossRef] [PubMed]
- Kubai, D.F.; Ris, H. Division in the dinoflagellate Gyrodinium cohnii (Schiller). A new type of nuclear reproduction. J. Cell Biol. 1969, 40, 508–528. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Brenner, S.; Cuzin, F. On the regulation of DNA replication in bacteria. Cold Spring Harb. Symp. Quant. Biol. 1963, 28, 329–348. [Google Scholar] [CrossRef]
- Oakley, B.R.; Dodge, J.D. Mitosis in the Cryptophyceae. Nature 1973, 244, 521–522. [Google Scholar] [CrossRef] [PubMed]
- Feder, N.; Sidman, R.L. Methods and principles of fixation by freeze-substitution. J. Cell Biol. 1958, 4, 593–600. [Google Scholar] [CrossRef]
- Heath, I.B. Mitosis in the fungus Thraustotheca clavata. J. Cell Biol. 1974, 60, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Ris, H.; Kubai, D.F. An unusual mitotic mechanism in the parasitic protozoan Syndinium sp. J. Cell Biol. 1974, 60, 702–720. [Google Scholar] [CrossRef] [PubMed]
- Kubai, D.F. The evolution of the mitotic spindle. Int. Rev. Cytol. 1975, 43, 167–227. [Google Scholar] [PubMed]
- He, X.; Asthana, S.; Sorger, P.K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 2000, 101, 763–775. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.D.; Tippit, D.H.; Leslie, R. Light and electron microscopic observations on cell division in two large pennate diatoms, Hantzschia and Nitzschia. I. Mitosis in vivo. Eur. J. Cell Biol. 1980, 21, 1–11. [Google Scholar] [PubMed]
- Nabeshima, K.; Nakagawa, T.; Straight, A.F.; Murray, A.; Chikashige, Y.; Yamashita, Y.M.; Hiraoka, Y.; Yanagida, M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 1998, 9, 3211–3225. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 1981, 84, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Fuge, H. The arrangement of microtubules and the attachment of chromosomes to the spindle during anaphase in tipulid spermatocytes. Chromosoma 1974, 45, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Metz, C.W. An apparent case of monocentric mitosis in Sciara (Diptera). Science 1926, 63, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Kubai, D.F. Meiosis in Sciara coprophila: Structure of the spindle and chromosome behavior during the first meiotic division. J. Cell Biol. 1982, 93, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Johnson, U.G.; Porter, K.R. Fine structure of cell division in Chlamydomonas reinhardi. Basal bodies and microtubules. J. Cell Biol. 1968, 38, 405–425. [Google Scholar] [CrossRef]
- Stafstrom, J.P.; Staehelin, L.A. Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo. Eur. J. Cell Biol. 1984, 34, 179–189. [Google Scholar] [PubMed]
- Oakley, B.R.; Dodge, J.D. Kinetochores associated with the nuclear envelope in the mitosis of a dinoflagellate. J. Cell Biol. 1974, 63, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Ritter, H.; Inoue, S.; Kubai, D. Mitosis in Barbulanympha. I. Spindle structure, formation, and kinetochore engagement. J. Cell Biol. 1978, 77, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Mazia, D.; Dan, K. The Isolation and Biochemical Characterization of the Mitotic Apparatus of Dividing Cells. Proc. Natl. Acad. Sci. USA 1952, 38, 825–835. [Google Scholar] [CrossRef]
- Mazia, D.; Mitchison, J.M.; Medina, H.; Harris, P. The direct isolation of the mitotic apparatus. J. Cell Biol. 1961, 10, 467–474. [Google Scholar] [CrossRef]
- Kane, R.E. The mitotic apparatus. Physical-chemical factors controlling stability. J. Cell Biol. 1965, 25, 137–144. [Google Scholar] [CrossRef]
- Forer, A. Characteristics of sea-urchin mitotic apparatus isolated using a dimethyl sulphoxide/glycerol medium. J. Cell Sci. 1974, 16, 481–497. [Google Scholar] [PubMed]
- Sakai, H.S.; Shimoda, S.; Hiramoto, Y. Mass isolation of mitotic apparatus using a glycerol/Mg2+/Triton X-100 medium. Exp. Cell Res. 1977, 104, 457–461. [Google Scholar] [CrossRef]
- Murphy, D.B. Identification of microtubule-associated proteins in the meiotic spindle of surf clam oocytes. J. Cell Biol. 1980, 84, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Schiff, P.B.; Fant, J.; Horwitz, S.B. Promotion of microtubule assembly in vitro by taxol. Nature 1979, 277, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Kuriyama, R.; Keryer, G.; Borisy, G.G. The mitotic spindle of Chinese hamster ovary cells isolated in taxol-containing medium. J. Cell Sci. 1984, 66, 265–275. [Google Scholar] [PubMed]
- Taylor, E.W. The Mechanism of Colchicine Inhibition of Mitosis. I. Kinetics of Inhibition and the Binding of H3-Colchicine. J. Cell Biol. 1965, 25, 145–160. [Google Scholar] [CrossRef]
- Borisy, G.G.; Taylor, E.W. The mechanism of action of colchicine. Binding of colchincine-3H to cellular protein. J. Cell Biol. 1967, 34, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Weisenberg, R.C. Microtubule formation in vitro in solutions containing low calcium concentrations. Science 1972, 177, 1104–1105. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann-Berling, H. Adenosinetriphosphate as the energy substance for cell movement. Biochim. Biophys. Acta 1954, 14, 182–194. [Google Scholar] [PubMed]
- Cande, W.Z.; Snyder, J.; Smith, D.; Summers, K.; McIntosh, J.R. A functional mitotic spindle prepared from mammalian cells in culture. Proc. Natl. Acad. Sci. USA 1974, 71, 1559–1563. [Google Scholar] [CrossRef] [PubMed]
- Rebhun, L.I.; Palazzo, R.E. In vitro reactivation of anaphase B in isolated spindles of the sea urchin egg. Cell Motil. Cytoskelet. 1988, 10, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Lohka, M.J.; Maller, J.L. Induction of nuclear envelope breakdown, chromosome condensation, and spindle formation in cell-free extracts. J. Cell Biol. 1988, 101, 518–523. [Google Scholar] [CrossRef]
- Lohka, M.J.; Hayes, M.K.; Maller, J.K. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc. Natl. Acad. Sci. USA 1988, 85, 3009–3013. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Pollard, T.D. Fluorescent antibody localization of myosin in the cytoplasm, cleavage furrow, and mitotic spindle of human cells. J. Cell Biol. 1976, 71, 848–875. [Google Scholar] [CrossRef] [PubMed]
- Izant, J.G.; Weatherbee, J.A.; McIntosh, J.R. A microtubule-associated protein in the mitotic spindle and the interphase nucleus. Nature 1982, 295, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Cande, W.Z.; Lazarides, E.; McIntosh, J.R. A comparison of the distribution of actin and tubulin in the mammalian mitotic spindle as seen by indirect immunofluorescence. J. Cell Biol. 1977, 72, 552–567. [Google Scholar] [CrossRef] [PubMed]
- Barak, L.S.; Nothnagel, E.A.; DeMarco, E.F.; Webb, W.W. Differential staining of actin in metaphase spindles with 7-nitrobenz-2-oxa-1,3-diazole-phallacidin and fluorescent DNase: Is actin involved in chromosomal movement? Proc. Natl. Acad. Sci. USA 1981, 78, 3034–3038. [Google Scholar] [CrossRef] [PubMed]
- Oakley, C.E.; Oakley, B.R. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature 1989, 338, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Pfarr, C.M.; Coue, M.; Grissom, P.M.; Hays, T.S.; Porter, M.E.; McIntosh, J.R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 1990, 345, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Steuer, E.R.; Wordeman, L.; Schroer, T.A.; Sheetz, M.P. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature 1990, 345, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Moroi, Y.; Peebles, C.; Fritzler, M.J.; Steigerwald, J.; Tan, E.M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl. Acad. Sci. USA 1980, 77, 1627–1631. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Halligan, N.; Cooke, C.; Rothfield, N. The kinetochore is part of the metaphase chromosome scaffold. J. Cell Biol. 1984, 98, 3352–3357. [Google Scholar] [CrossRef]
- Bernat, R.L.; Borisy, G.G.; Rothfield, N.F.; Earnshaw, W.C. Injection of anticentromere antibodies in interphase disrupts events required for chromosome movement at mitosis. J. Cell Biol. 1990, 111, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.K.; O’Day, K.; Trong, H.L.; Charbonneau, H.; Margolis, R.L. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 1991, 88, 3734–3738. [Google Scholar] [CrossRef] [PubMed]
- Wald, H. Cytologic studies of the abnormal development of the eggs of the Claret mutant type of Drosophila simulans. Genetics 1935, 21, 264–281. [Google Scholar]
- Davis, D.G. Chromosome Behavior under the Influence of Claret-Nondisjunctional in Drosophila melanogaster. Genetics 1969, 61, 577–594. [Google Scholar] [PubMed]
- Walker, R.A.; Salmon, E.D.; Endow, S.A. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature 1990, 347, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Ostergren, G. Description of unpublished work on mitotic mutants in Lily, given to the author during a meeting at the Swedish. University of Agricultural Sciences in Uppsala: Sweden, 1976. [Google Scholar]
- Baker, B.S. Paternal loss (pal): A meiotic mutant in Drosophila melanogaster causing loss of paternal chromosomes. Genetics 1975, 80, 267–296. [Google Scholar] [PubMed]
- Gatti, M.; Baker, B.S. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989, 3, 438–453. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.; Culotti, J.; Ried, B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 1962, 66, 352–359. [Google Scholar] [CrossRef]
- Nurse, P. Genetic control of cell size at cell division in yeast. Nature 1975, 256, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Morris, N.R. Mitotic mutants of Aspergillus nidulans. Genet. Res. 1975, 26, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Funahashi, S.; Uemura, T.; Yanagida, M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986, 5, 2973–2979. [Google Scholar] [PubMed]
- Hoyt, M.A.; Stearns, T.; Botstein, D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol. Cell. Biol. 1990, 10, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.T.; Saxton, W.M.; Goldstein, L.S. Isolation and characterization of the gene encoding the heavy chain of Drosophila kinesin. Proc. Natl. Acad. Sci. USA 1988, 85, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Murray, A.W. Feedback control of mitosis in budding yeast. Cell 1991, 66, 519–531. [Google Scholar] [CrossRef]
- Hoyt, M.A.; He, L.; Loo, K.K.; Saunders, W.S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 1992, 118, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Rout, M.P.; Kilmartin, J.V. Components of the yeast spindle and spindle pole body. J. Cell Biol. 1990, 111, 1913–1927. [Google Scholar] [CrossRef] [PubMed]
- Schrader, F. On the reality of spindle fibers. Biol. Bull. 1934, 67, 519–533. [Google Scholar] [CrossRef]
- Carlson, J.G. Mitotic Behavior of Induced Chromosomal Fragments Lacking Spindle Attachments in the Neuroblasts of the Grasshopper. Proc. Natl. Acad. Sci. USA 1938, 24, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Hughes-Schrader, S.; Ris, H. The diffuse spindle attachment of coccids, verified by the mitotic behavior of induced chromosome fragments. J. Exp. Zool. 1941, 87, 429–456. [Google Scholar] [CrossRef]
- White, M.J.D. The Effect of X-Rays on the First Meiotic Division in Three Species of Orthoptera. Proc. R. Soc. Lond. Ser. B 1937, 124, 183–196. [Google Scholar] [CrossRef]
- Ris, H. A quantitative study of anaphase movement in the aphid Tamalia. Biol. Bull. 1943, 85, 164–178. [Google Scholar] [CrossRef]
- Brinkley, B.R.; Stubblefield, E.; Hsu, T.C. The effects of colcemid inhibition and reversal on the fine structure of the mitotic apparatus of Chinese hamster cells in vitro. J. Ultrastruct. Res. 1967, 19, 1–18. [Google Scholar] [CrossRef]
- Belar, K. Beiträge zur Kausalanalyse der Mitose. Roux Arch Entw Mech Org 1929, 118, 359–484. [Google Scholar] [CrossRef]
- Dietz, R. Multiple Geschlechchromosomen bei den cypriden Ostracoden, ihre Evolution and ihr Teilungsverhalten. Chromosoma 1958, 9, 359–440. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. Recurrent pole-to-pole movements of the sex chromosome during prometaphase I in Melanoplus differentialis spermatocytes. Chromosoma 1961, 12, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. Chromosome Velocity during Mitosis as a Function of Chromosome Size and Position. J. Cell Biol. 1965, 25, 119–135. [Google Scholar] [CrossRef]
- Hughes-Schrader, S. Polarization, kinetochore movements, and bivalent structure in the meiosis of male mantids. Biol. Bull. 1943, 85, 265–300. [Google Scholar] [CrossRef]
- Hughes-Schrader, S. The “Pre-Metaphase Stretch” and kinetochore orientation in Phasmids. Chromosoma 1946, 3, 1–21. [Google Scholar] [CrossRef]
- Shimamura, T. On the mechanism of nuclear division and chromosome arrangement. VI. Studies on the effect of the centrifugal force upon nuclear division. Cytologia 1940, 11, 186–216. [Google Scholar]
- Beams, H.W.; King, R.L. The effect of ultracentrifuging upon chick embryonic cells, with special reference to the “resting” nucleus and the mitotic spindle. Biol. Bull. 1936, 71, 188–198. [Google Scholar] [CrossRef]
- Inoue, S.; Sato, H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 1967, 50, 259–292. [Google Scholar] [CrossRef]
- Rebhun, L.I.; Sawada, N. Augmentation and dispersion of the in vivo mitotic apparatus of living marine eggs. Protoplasma 1969, 68, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S. The effect of colchicine on the microscopic and submicroscopic structure of the mitotic spindle. Exp. Cell Res. Suppl. 1952, 2, 305–318. [Google Scholar]
- Taylor, E.W. Dynamics of Spindle Formation and its Inhibition by Chemicals. J. Cell Biol. 1959, 6, 193–196. [Google Scholar] [CrossRef]
- Taylor, E.W. Relation of Protein Synthesis to the Division Cycle in Mammalian Cell Cultures. J. Cell Biol. 1963, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Salmon, E.D.; Begg, D.A. Functional implications of cold-stable microtubules in kinetochore fibers of insect spermatocytes during anaphase. J. Cell Biol. 1980, 85, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Salmon, E.D. Spindle microtubules: Thermodynamics of in vivo assembly and role in chromosome movement. Ann. N. Y. Acad. Sci. 1975, 253, 383–406. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Ellis, G.W.; Inoue, S. Microtubular origin of mitotic spindle form birefringence. Demonstration of the applicability of Wiener’s equation. J. Cell Biol. 1975, 67, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S. Microtubule dynamics and chromosome motion. In Cell Motility; Cold Spring Harbor Conferences on Cell Proliferation; Cold Spring Harbor Press: Cold Spring Harbor, NY, USA, 1976; Volume 3, pp. 1317–1328. [Google Scholar]
- Forer, A. Characterization of the mitotic traction system, and evidence that birefringent spindle fibers neither produce nor transmit force for chromosome movement. Chromosoma 1966, 19, 44–98. [Google Scholar] [CrossRef] [PubMed]
- Leslie, R.J.; Pickett-Heaps, J.D. Ultraviolet microbeam irradiations of mitotic diatoms: Investigation of spindle elongation. J. Cell Biol. 1983, 96, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.J.; Forer, A. Ultraviolet microbeam irradiation of chromosomal spindle fibres shears microtubules and permits study of the new free ends in vivo. J. Cell Sci. 1988, 91, 455–468. [Google Scholar] [PubMed]
- McDonald, K.L.; Edwards, M.K.; McIntosh, J.R. Cross-sectional structure of the central mitotic spindle of Diatoma vulgare. Evidence for specific interactions between antiparallel microtubules. J. Cell Biol. 1979, 83, 443–461. [Google Scholar] [CrossRef] [PubMed]
- Scholey, J.M.; Civelekoglu-Scholey, G.; Brust-Mascher, I.; Anaphase, B. Biology 2017. [CrossRef]
- Salmon, E.D.; McKeel, M.; Hays, T. Rapid rate of tubulin dissociation from microtubules in the mitotic spindle in vivo measured by blocking polymerization with colchicine. J. Cell Biol. 1984, 99, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.; Kirschner, M. Dynamic instability of microtubule growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Leslie, R.J.; Saxton, W.M.; Mitchison, T.J.; Neighbors, B.; Salmon, E.D.; McIntosh, J.R. Assembly properties of fluorescein-labeled tubulin in vitro before and after fluorescence bleaching. J. Cell Biol. 1984, 99, 2146–2156. [Google Scholar] [CrossRef] [PubMed]
- Gorbsky, G.J.; Sammak, P.J.; Borisy, G.G. Chromosomes move poleward in anaphase along stationary microtubules that coordinately disassemble from their kinetochore ends. J. Cell Biol. 1987, 104, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Salmon, E.D.; Leslie, R.J.; Saxton, W.M.; Karow, M.L.; McIntosh, J.R. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 1984, 99, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, P.; Salmon, E.D. Analysis of the treadmilling model during metaphase of mitosis using fluorescence redistribution after photobleaching. J. Cell Biol. 1986, 102, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Cassimeris, L.; Inoue, S.; Salmon, E.D. Microtubule dynamics in the chromosomal spindle fiber: Analysis by fluorescence and high-resolution polarization microscopy. Cell Motil. Cytoskelet. 1988, 10, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.; Evans, L.; Schulze, E.; Kirschner, M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 1986, 45, 515–527. [Google Scholar] [CrossRef]
- Mitchison, T.J. Polewards microtubule flux in the mitotic spindle: Evidence from photoactivation of fluorescence. J. Cell Biol. 1989, 109, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B.; Staehly, C.A. Chromosome micromanipulation. I. The mechanics of chromosome attachment to the spindle. Chromosoma 1967, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Begg, D.A.; Ellis, G.W. Micromanipulation studies of chromosome movement. II. Birefringent chromosomal fibers and the mechanical attachment of chromosomes to the spindle. J. Cell Biol. 1979, 82, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B.; Koch, C.A. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 1969, 43, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. Chromosome micromanipulation. II. Induced reorientation and the experimental control of segregation in meiosis. Chromosoma 1967, 21, 17–50. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 1982, 79, 1–58. [Google Scholar] [PubMed]
- Henderson, S.A.; Koch, C.A. Co-orientation stability by physical tension: A demonstration with experimentally interlocked bivalents. Chromosoma 1970, 29, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B.; Kubai, D.F. Microtubules, chromosome movement, and reorientation after chromosomes are detached from the spindle by micromanipulation. Chromosoma 1985, 92, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ault, J.G.; Nicklas, R.B. Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma 1989, 98, 33–39. [Google Scholar] [CrossRef] [PubMed]
- McNeill, P.A.; Berns, M.W. Chromosome behavior after laser microirradiation of a single kinetochore in mitotic PtK2 cells. J. Cell Biol. 1981, 88, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.W. Brownian and saltatory movements of cytoplasmic granules and the movement of anaphase chromosomes. In Proceedings of the Fourth International Congress on Rheology; Copley, A.L., Ed.; NASA: New York, NY, USA, 1965; Part 4; pp. 175–191. [Google Scholar]
- Nicklas, R.B. Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 1983, 97, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. Chromosome movement: Current models and experiments on living cells. In “Molecules and Cell Movement”; lnoué, S., Stephens, R.E., Eds.; Raven Press: Raven, New York, NY, USA, 1975; pp. 97–118. [Google Scholar]
- Forer, A. Possible roles of microtubules and actin-like filaments during cell-division. In Cell Cycle Controls; Academic Press: New York, NY, USA, 1974; pp. 319–336. [Google Scholar]
- Rashevsky, N. Some remarks on the movement of chromosomes during cell division. Bull. Math. Biophys. 1941, 3, 1–3. [Google Scholar] [CrossRef]
- Ostergren, G. Equilibrium of trivalents and the mechanism of chromosome movement. Hereditas 1945, 31, 498–511. [Google Scholar]
- Ostergren, G. Considerations on some elementary features of mitosis. Hereditas 1950, 36, 1–19. [Google Scholar] [CrossRef]
- Wise, D. On the mechanism of prometaphase congression: Chromosome velocity as a function of position on the spindle. Chromosoma 1978, 69, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Wada, B.; Izutsu, K. Effects of ultraviolet microbeam irradiations on mitosis studied in Tradescantia cells in vivo. Cytologia 1961, 26, 480–491. [Google Scholar] [CrossRef]
- Ostergren, G. The mechanism of co-ordination of bivalents and multivalents. Hereditas 1951, 37, 85–156. [Google Scholar] [CrossRef]
- Hays, T.S.; Wise, D.; Salmon, E.D. Traction force on a kinetochore at metaphase acts as a linear function of kinetochore fiber length. J. Cell Biol. 1982, 93, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Hays, T.S.; Salmon, E.D. Poleward force at the kinetochore in metaphase depends on the number of kinetochore microtubules. J. Cell Biol. 1990, 110, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L.; Davison, E.A.; Jensen, L.C.; Cassimeris, L.; Salmon, E.D. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 1986, 103, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Darlington, C.D. Recent Advances in Cytology; Blankiston's Son & Co. Inc: Philadelphia, PA, USA, 1937. [Google Scholar]
- Cornman, I. A summary of evidence in favor of the traction fiber in mitosis. Am. Nat. 1944, 78, 410–422. [Google Scholar] [CrossRef]
- Oosawa, F.; Kasai, M. A theory of linear and helical aggregations of macromolecules. J. Mol. Biol. 1962, 4, 10–21. [Google Scholar] [CrossRef]
- Forer, A. Local Reduction of Spindle Fiber Birefringence in Living Nephrotoma Suturalis (Loew) Spermatocytes Induced by Ultraviolet Microbeam Irradiation. J. Cell Biol. 1965, 25, 95–117. [Google Scholar] [CrossRef]
- Subirana, J.A. Role of spindle microtubules in mitosis. J. Theor. Biol. 1968, 20, 117–123. [Google Scholar] [CrossRef]
- Schibler, M.J.; Pickett-Heaps, J.D. Mitosis in Oedogonium: Spindle microfilaments and the origin of the kinetochore fiber. Eur. J. Cell Biol. 1980, 22, 687–698. [Google Scholar] [PubMed]
- Field, C.M.; Lénárt, P. Bulk cytoplasmic actin and its functions in meiosis and mitosis. Curr. Biol. 2011, 21, R825–R830. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, I.R.; Rowe, A.J. Dynein: A Protein with Adenosine Triphosphatase Activity from Cilia. Science 1965, 149, 424–426. [Google Scholar] [CrossRef] [PubMed]
- Euteneuer, U.; McIntosh, J.R. Structural polarity of kinetochore microtubules in PtK1 cells. J. Cell Biol. 1981, 89, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Mazia, D. The Cell in Mitosis. In The Cell; Academic Press: New York, NY, USA, 1961; Volume 3. [Google Scholar]
- Margolis, R.L.; Wilson, L.; Keifer, B.I. Mitotic mechanism based on intrinsic microtubule behaviour. Nature 1978, 272, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.; Mitchison, T. Beyond self-assembly: From microtubules to morphogenesis. Cell 1986, 45, 329–342. [Google Scholar] [CrossRef]
- Holy, T.E.; Leibler, S. Dynamic instability of microtubules as an efficient way to search in space. Proc. Natl. Acad. Sci. USA 1994, 91, 5682–5685. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Wollman, R.; Silkworth, W.T.; Nardi, I.K.; Cimini, D.; Mogilner, A. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proc. Natl. Acad. Sci. USA 2009, 106, 15708–15713. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.L.; Kirschner, M.W. Bioenergetics and kinetics of microtubule and actin filament assembly-disassembly. Int. Rev. Cytol. 1982, 78, 1–125. [Google Scholar] [PubMed]
- Hill, T.L. Theoretical problems related to the attachment of microtubules to kinetochores. Proc. Natl. Acad. Sci. USA 1985, 82, 4404–4408. [Google Scholar] [CrossRef] [PubMed]
- Koshland, D.E.; Mitchison, T.J.; Kirschner, M.W. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature 1988, 331, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Coue, M.; Lombillo, V.A.; McIntosh, J.R. Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 1991, 112, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Brinkley, B.R.; Cartwright, J. Ultrastructural analysis of mitotic spindle elongation in mammalian cells in vitro. Direct microtubule counts. J. Cell Biol. 1971, 50, 416–431. [Google Scholar] [CrossRef] [PubMed]
- Tippit, D.H.; Fields, C.T.; O’Donnell, K.L.; Pickett-Heaps, J.D.; McLaughlin, D.J. The organization of microtubules during anaphase and telophase spindle elongation in the rust fungus Puccinia. Eur. J. Cell Biol. 1984, 34, 34–44. [Google Scholar] [PubMed]
- Masuda, H.; Hirano, T.; Yanagida, M.; Cande, W.Z. In vitro reactivation of spindle elongation in fission yeast nuc2 mutant cells. J. Cell Biol. 1990, 110, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.K.; Odde, D.J. Modeling of chromosome motility during mitosis. Curr. Opin. Cell Biol. 2006, 18, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Nedelec, F.J.; Surrey, T.; Maggs, A.C.; Leibler, S. Self-organization of microtubules and motors. Nature 1997, 389, 305–308. [Google Scholar] [PubMed]








© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McIntosh, J.R.; Hays, T. A Brief History of Research on Mitotic Mechanisms. Biology 2016, 5, 55. https://doi.org/10.3390/biology5040055
McIntosh JR, Hays T. A Brief History of Research on Mitotic Mechanisms. Biology. 2016; 5(4):55. https://doi.org/10.3390/biology5040055
Chicago/Turabian StyleMcIntosh, J. Richard, and Thomas Hays. 2016. "A Brief History of Research on Mitotic Mechanisms" Biology 5, no. 4: 55. https://doi.org/10.3390/biology5040055





