Applications of Microalgal Biotechnology for Disease Control in Aquaculture
Abstract
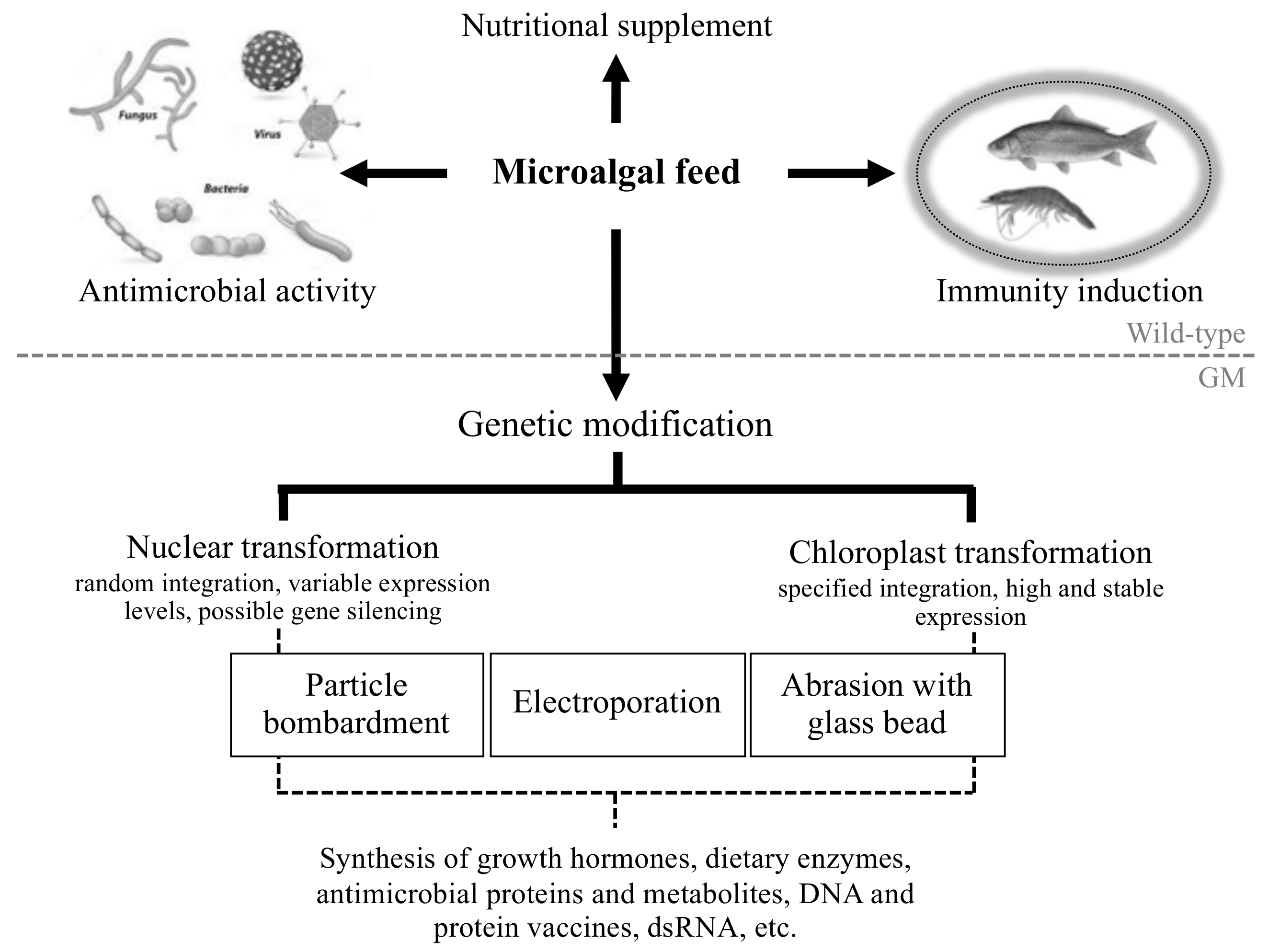
:1. Introduction
2. The Application of Microalgae in Aquaculture
3. Microalgae as a Health-Promoting Supplement for Coping with Aquaculture Disease
4. Engineering Microalgae to Produce Novel Antiviral and Antibacterial Biomolecules
5. Challenges and Constraints in Algal Research
5.1. Further Advances in Microalgal Genetic Engineering
5.2. Large-Scale Production and Downstream Processing
5.3. Regulation, Risk, and Public Acceptance
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- The Food and Agriculture Organization of the United Nations (FAO). The State of World Fisheries and Aquaculture 2016. Contributing to Food Security and Nutrition for All; FAO: Rome, Italy, 2016. [Google Scholar]
- Bondad-Reantaso, M.G.; Subasinghe, R.P.; Arthur, J.R.; Ogawa, K.; Chinabut, S.; Adlard, R.; Tan, Z.; Shariff, M. Disease and health management in Asian aquaculture. Vet. Parasitol. 2005, 132, 249–272. [Google Scholar] [CrossRef] [PubMed]
- Kusumaningrum, H.P.; Zainuri, M. Detection of bacteria and fungi associated with Penaeus monodon postlarvae mortality. Procedia Environ. Sci. 2015, 23, 329–337. [Google Scholar] [CrossRef]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P.; Salachan, P.V.; Sritunyalucksana, K.; Flegel, T.W.; Itsathitphaisarn, O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 2016, 452, 69–87. [Google Scholar] [CrossRef]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens; Disease of Farmed and Wild Fish, 5th ed.; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Crane, M.; Hyatt, A. Viruses of fish: An overview of significant pathogens. Viruses 2011, 3, 2025–2046. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, K.; Shariff, M.; Omar, A.R.; Hair-Bejo, M. Megalocytivirus infection in fish. Rev. Aquac. 2012, 4, 221–233. [Google Scholar] [CrossRef]
- Defoirdt, T.; Sorgeloos, P.; Bossier, P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr. Opin. Microbiol. 2011, 14, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016, 16, 127–133. [Google Scholar] [CrossRef]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The rising tide of antimicrobial Resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef] [PubMed]
- Plant, K.P.; Lapatra, S.E. Advances in fish vaccine delivery. Dev. Comp. Immunol. 2011, 35, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Dhar, A.; Allnutt, F. Challenges and opportunities in developing oral vaccines against viral diseases of fish. J. Mar. Sci. Res. Dev. S 2011, 1–6. [Google Scholar] [CrossRef]
- Kibenge, F.S.; Godoy, M.G.; Fast, M.; Workenhe, S.; Kibenge, M.J. Countermeasures against viral diseases of farmed fish. Antivir. Res. 2012, 95, 257–281. [Google Scholar] [CrossRef] [PubMed]
- Spolaore, P.; Joannis-Cassa, C.; Duran, E.; Isambert, A. Commercial applications of microalgae. J. Biosci. Bioeng. 2006, 101, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, M.R.; Lutzu, G.A.; Alam, A.; Sarker, P.; Chowdhury, M.A.K.; Parsaeimehr, A.; Liang, Y.; Daroch, M. Microalgae in aquafeeds for a sustainable aquaculture industry. J. Appl. Phycol. 2018, 30, 197–213. [Google Scholar] [CrossRef]
- Falaise, C.; François, C.; Travers, M.-A.; Morga, B.; Haure, J.; Tremblay, R.; Turcotte, F.; Pasetto, P.; Gastineau, R.; Hardivillier, Y.; et al. Antimicrobial compounds from Eukaryotic Microalgae against human pathogens and diseases in aquaculture. Mar. Drugs 2016, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Lin, H.; Jiang, P. Advances in genetic engineering of marine algae. Biotechnol. Adv. 2012, 30, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Spicer, A.; Purton, S. Genetic engineering of microalgae: Current status and future prospects. In Microalgal Production for Biomass and High-Value Products; Slocombe, S.P., Benemann, J.R., Eds.; CRC Press: Boca Raton, FL, USA, 2016; pp. 139–164. [Google Scholar]
- Taunt, H.N.; Stoffels, L.; Purton, S. Green biologics: The algal chloroplast as a platform for making biopharmaceuticals. Bioengineered 2018, 9, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Dang, V.T.; Li, Y.; Speck, P.; Benkendorff, K. Effects of micro and macroalgal diet supplementations on growth and immunity of greenlip abalone, Haliotis laevigata. Aquaculture 2011, 320, 91–98. [Google Scholar] [CrossRef]
- Yaakob, Z.; Ali, E.; Zainal, A.; Mohamad, M.; Takriff, M.S. An overview: Biomolecules from microalgae for animal feed and aquaculture. J. Biol. Res.-Thessalon. 2014, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Austin, B.; Day, J. Inhibition of prawn pathogenic Vibrio spp. by a commercial spray-dried preparation of Tetraselmis suecica. Aquaculture 1990, 90, 389–392. [Google Scholar] [CrossRef]
- Viso, A.; Pesando, D.; Baby, C. Antibacterial and antifungal properties of some marine diatoms in culture. Bot. Mar. 1987, 30, 41–46. [Google Scholar] [CrossRef]
- Das, B.; Pradhan, J. Antibacterial properties of selected freshwater microalgae against pathogenic bacteria. Indian J. Fish. 2010, 57, 61–66. [Google Scholar]
- Natrah, F.; Kenmegne, M.M.; Wiyoto, W.; Sorgeloos, P.; Bossier, P.; Defoirdt, T. Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquaculture 2011, 317, 53–57. [Google Scholar] [CrossRef]
- Benkendorff, K.; Davis, A.R.; Rogers, C.N.; Bremner, J.B. Free fatty acids and sterols in the benthic spawn of aquatic molluscs, and their associated antimicrobial properties. J. Exp. Mar. Biol. Ecol. 2005, 316, 29–44. [Google Scholar] [CrossRef]
- Desbois, A.P.; Mearns-Spragg, A.; Smith, V.J. A fatty acid from the diatom Phaeodactylum tricornutum is antibacterial against diverse bacteria including multi-resistant Staphylococcus aureus (MRSA). Mar. Biotechnol. 2009, 11, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Nonwachai, T.; Purivirojkul, W.; Limsuwan, C.; Chuchird, N.; Velasco, M.; Dhar, A.K. Growth, nonspecific immune characteristics, and survival upon challenge with Vibrio harveyi in Pacific white shrimp (Litopenaeus vannamei) raised on diets containing algal meal. Fish Shellfish Immunol. 2010, 29, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Merchie, G.; Kontara, E.; Lavens, P.; Robles, R.; Kurmaly, K.; Sorgeloos, P. Effect of vitamin C and astaxanthin on stress and disease resistance of postlarval tiger shrimp, Penaeus monodon (Fabricius). Aquac. Res. 1998, 29, 579–585. [Google Scholar] [CrossRef]
- Babin, A.; Biard, C.; Moret, Y. Dietary supplementation with carotenoids improves immunity without increasing its cost in a crustacean. Am. Nat. 2010, 176, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Kontara, E.; Merchie, G.; Lavens, P.; Robles, R.; Nelis, H.; De Leenheer, A.; Sorgeloos, P. Improved production of postlarval white shrimp through supplementation of L-ascorbyl-2-polyphosphate in their diet. Aquac. Int. 1997, 5, 127–136. [Google Scholar]
- Kanazawa, A. Recent developments in shrimp nutrition and feed industry. In INDAQUA ’95 Exposition of Indian Aquaculture, Madras, 27–30 January 1995; The Marine Products Export Development Agency: Cochin, India, 1995; pp. 28–29. [Google Scholar]
- Radhakrishnan, S.; Bhavan, P.S.; Seenivasan, C.; Shanthi, R.; Muralisankar, T. Replacement of fishmeal with Spirulina platensis, Chlorella vulgaris and Azolla pinnata on non-enzymatic and enzymatic antioxidant activities of Macrobrachium rosenbergii. J. Basic Appl. Zool. 2014, 67, 25–33. [Google Scholar] [CrossRef]
- Tayag, C.M.; Lin, Y.C.; Li, C.C.; Liou, C.H.; Chen, J.C. Administration of the hot-water extract of Spirulina platensis enhanced the immune response of white shrimp Litopenaeus vannamei and its resistance against Vibrio alginolyticus. Fish Shellfish Immunol. 2010, 28, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Newaj-Fyzul, A.; Austin, B. Probiotics, immunostimulants, plant products and oral vaccines, and their role as feed supplements in the control of bacterial fish diseases. J. Fish Dis. 2015, 38, 937–955. [Google Scholar] [CrossRef] [PubMed]
- Debtanu Barman, P.N.; Mandal, S.C.; Kumar, V. Immunostimulants for Aquaculture Health Management. J. Mar. Sci. Res. Dev. 2013, 3. [Google Scholar] [CrossRef]
- Carton-Kawagoshi, R.J.; Caipang, C.M. Algal-derived products and their role in shrimp immunity. In Biotechnological Advances in Shrimp Health Management in the Philippines; Caipang, C.M., Bacano-Maningas, M.B., Fagutao, F.F., Eds.; Research Signpost: Trivandrum, India, 2015; pp. 73–88. [Google Scholar]
- Wang, H.; Dai, A.; Liu, F.; Guan, Y. Effects of dietary astaxanthin on the immune response, resistance to white spot syndrome virus and transcription of antioxidant enzyme genes in Pacific white shrimp Litopenaeus vannamei. Iran. J. Fish. Sci. 2015, 14, 699–718. [Google Scholar]
- Supamattaya, K.; Kiriratnikom, S.; Boonyaratpalin, M.; Borowitzka, L. Effect of a Dunaliella extract on growth performance, health condition, immune response and disease resistance in black tiger shrimp (Penaeus monodon). Aquaculture 2005, 248, 207–216. [Google Scholar] [CrossRef]
- Madhumathi, M.; Rengasamy, R. Antioxidant status of Penaeus monodon fed with Dunaliella salina supplemented diet and resistance against WSSV. Int. J. Eng. Sci. Technol. 2011, 3, 7249–7259. [Google Scholar]
- Doron, L.; Segal, N.A.; Shapira, M. Transgene expression in microalgae—From tools to applications. Front. Plant Sci. 2016, 7, 505. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Fan, C.; Chen, Y.; Hu, Z. The potential for microalgae as bioreactors to produce pharmaceuticals. Int. J. Mol. Sci. 2016, 17, 962. [Google Scholar] [CrossRef] [PubMed]
- Scaife, M.A.; Nguyen, G.T.D.T.; Rico, J.; Lambert, D.; Helliwell, K.E.; Smith, A.G. Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant J. 2015, 82, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Dyo, Y.M.; Purton, S. The algal chloroplast as a synthetic biology platform for production of therapeutic proteins. Microbiology 2018, 164, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Economou, C.; Wannathong, T.; Szaub, J.; Purton, S. A simple, low-cost method for chloroplast transformation of the green alga Chlamydomonas reinhardtii. Methods Mol. Biol. 2014, 1132, 401–411. [Google Scholar]
- Rathod, J.P.; Gade, R.M.; Rathod, D.R.; Dudhare, M. A review on molecular tools of microalgal genetic transformation and their application for overexpression of different genes. Int. J. Curr. Microbiol. App. Sci. 2017, 6, 3191–3207. [Google Scholar] [CrossRef]
- Jeon, S.; Lim, J.; Lee, H.; Shin, S.; Kang, N.; Park, Y.I.; Oh, H.-M.; Jeong, W.-J.; Jeong, B.; Chang, Y.K. Current status and perspectives of genome editing technology for microalgae. Biotechnol. Biofuels 2017, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Wannathong, T.; Waterhouse, J.C.; Young, R.E.; Economou, C.K.; Purton, S. New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2016, 100, 5467–5477. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Li, X.; Xu, Z.; Qi, J. Dunaliella salina as a novel host for the production of recombinant proteins. Appl. Microbiol. Biotechnol. 2014, 98, 4293–4300. [Google Scholar] [CrossRef] [PubMed]
- Scranton, M.A.; Ostrand, J.T.; Georgianna, D.R.; Lofgren, S.M.; Li, D.; Ellis, R.C.; Carruthers, D.N.; Dräger, A.; Masica, D.L.; Mayfield, S.P. Synthetic promoters capable of driving robust nuclear gene expression in the green alga Chlamydomonas reinhardtii. Algal Res. 2016, 15, 135–142. [Google Scholar] [CrossRef]
- Rasala, B.A.; Muto, M.; Sullivan, J.; Mayfield, S.P. Improved heterologous protein expression in the chloroplast of Chlamydomonas reinhardtii through promoter and 5′ untranslated region optimization. Plant Biotechnol. J. 2011, 9, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Tsai, H.J. Transgenic microalgae as a non-antibiotic bactericide producer to defend against bacterial pathogen infection in the fish digestive tract. Fish Shellfish Immunol. 2009, 26, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Feng, W.; Zhao, L.; Gu, H.; Li, Q.; Shi, K.; Guo, S.; Zhang, N. Preparation of transgenic Dunaliella salina for immunization against white spot syndrome virus in crayfish. Arch. Virol. 2014, 159, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Unajak, S.; Kiataramgul, A.; Wannathong, T.W.; Mavichak, R.; Yokthongwattana, C.; Areechon, N. Development of Chlamydomonas reinhardtii for control white spot syndrome virus in shrimp (Penaeus vannamei). In Proceedings of the Asian-Pacific Aquaculture 2016 Conference, Surabaya, Indonesia, 26–29 April 2016; p. 610. [Google Scholar]
- Siripornadulsil, S.; Dabrowski, K.; Sayre, R. Microalgal vaccines. Adv. Exp. Med. Biol. 2007, 616, 122–128. [Google Scholar] [PubMed]
- Surzycki, R.; Greenham, K.; Kitayama, K.; Dibal, F.; Wagner, R.; Rochaix, J.D.; Ajam, T.; Surzycki, S. Factors effecting expression of vaccines in microalgae. Biologicals 2009, 37, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt-Clermont, M. Fischimpfung mit Chlamydomonas, die Bakterielle Antigene im Chloroplast Exprimieren. Available online: https://tinyurl.com/ydhzydbu (accessed on 11 April 2018).
- Michelet, L.; Lefebvre-Legendre, L.; Burr, S.E.; Rochaix, J.D.; Goldschmidt-Clermont, M. Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol. J. 2011, 9, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Somchai, P.; Jitrakorn, S.; Thitamadee, S.; Meetam, M.; Saksmerprome, V. Use of microalgae Chlamydomonas reinhardtii for production of double-stranded RNA against shrimp virus. Aquac. Rep. 2016, 3, 178–183. [Google Scholar] [CrossRef]
- Saksmerprome, V.; Charoonnart, P.; Gangnonngiw, W.; Withyachumnarnkul, B. A novel and inexpensive application of RNAi technology to protect shrimp from viral disease. J. Virol. Methods 2009, 162, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Thammasorn, T.; Sangsuriya, P.; Meemetta, W.; Senapin, S.; Jitrakorn, S.; Rattanarojpong, T.; Saksmerprome, V. Large-scale production and antiviral efficacy of multi-target double-stranded RNA for the prevention of white spot syndrome virus (WSSV) in shrimp. BMC Biotechnol. 2015, 15, 110. [Google Scholar] [CrossRef] [PubMed]
- Saksmerprome, V.; Thammasorn, T.; Jitrakorn, S.; Wongtripop, S.; Borwornpinyo, S.; Withyachumnarnkul, B. Using double-stranded RNA for the control of Laem-Singh Virus (LSNV) in Thai P. monodon. J. Biotechnol. 2013, 164, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Wang, S.; Ou, R.; Samrakandi, M.; Beerntsen, B.T.; Sayre, R.T. Development of an RNAi based microalgal larvicide to control mosquitoes. Malar. World J. 2013, 4, 1–7. [Google Scholar]
- Gumpel, N.J.; Purton, S. Playing tag with Chlamydomonas. Trends Cell Biol. 1994, 4, 299–301. [Google Scholar] [CrossRef]
- Bertalan, I.; Munder, M.C.; Weiß, C.; Kopf, J.; Fischer, D.; Johanningmeier, U. A rapid, modular and marker-free chloroplast expression system for the green alga Chlamydomonas reinhardtii. J. Biotechnol. 2015, 195, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Charoonnart, P. National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Thailand. Unpublished work. 2018. [Google Scholar]
- Scaife, M.A.; Smith, A.G. Towards developing algal synthetic biology. Biochem. Soc. Trans. 2016, 44, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.R.; Purton, S. Genetic engineering of eukaryotic algae: Progress and prospects. J. Phycol. 1997, 3, 713–722. [Google Scholar] [CrossRef]
- Ferenczi, A.; Pyott, D.E.; Xipnitou, A.; Molnar, A. Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc. Natl. Acad. Sci. USA 2017, 114, 13567–13572. [Google Scholar] [CrossRef] [PubMed]
- Slattery, S.S.; Diamond, A.; Wang, H.; Therrien, J.A.; Lant, J.T.; Jazey, T.; Lee, K.; Klassen, Z.; Desgagné-Penix, I.; Karas, B.J.; et al. An expanded plasmid-based genetic toolbox enables Cas9 genome editing and stable maintenance of synthetic pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 2018, 7, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Daboussi, F. Genetic and metabolic engineering in diatoms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef] [PubMed]
- Potvin, G.; Zhang, Z. Strategies for high-level recombinant protein expression in transgenic microalgae: A review. Biotechnol. Adv. 2010, 28, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Chung, H.; Kim, T. Codon-optimized expression of fish iridovirus capsid protein in yeast and its application as an oral vaccine candidate. J. Fish Dis. 2013, 36, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Kanesaki, Y.; Hirooka, S.; Era, A.; Sumiya, N.; Yoshikawa, H.; Tanaka, K.; Miyagishima, S.Y. A nitrogen source-dependent inducible and repressible gene expression system in the red alga Cyanidioschyzon merolae. Front. Plant Sci. 2015, 6, 657. [Google Scholar] [CrossRef] [PubMed]
- Iwai, M.; Hori, K.; Sasaki-Sekimoto, Y.; Shimojima, M.; Ohta, H. Manipulation of oil synthesis in Nannochloropsis strain NIES-2145 with a phosphorus starvation-inducible promoter from Chlamydomonas reinhardtii. Front. Microbiol. 2015, 6, 912. [Google Scholar] [CrossRef] [PubMed]
- Stoffels, L.; Taunt, H.N.; Charalambous, B.; Purton, S. Synthesis of bacteriophage lytic proteins against Streptococcus pneumoniae in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 2017, 15, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Kalatzis, P.G.; Castillo, D.; Katharios, P.; Middelboe, M. Bacteriophage interactions with marine pathogenic Vibrios: Implications for phage therapy. Antibiotics 2018, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Li, S.S.; Huang, R.; Tsai, H.J. Conditional production of a functional fish growth hormone in the transgenic line of Nannochloropsis oculata (Eustigmatophyceae). J. Phycol. 2008, 44, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, Y.T.; Cho, J.J.; Bae, J.H.; Hur, S.B.; Hwang, I.; Choi, T.J. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol. 2002, 4, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Kwon, T.; Seo, J.; Kim, T. Oral immunization of fish against iridovirus infection using recombinant antigen produced from rice callus. Vaccine 2013, 31, 5210–5215. [Google Scholar] [CrossRef] [PubMed]
- Allnutt, F.C.; Bowers, R.M.; Rowe, C.G.; Vakharia, V.N.; LaPatra, S.E.; Dhar, A.K. Antigenicity of infectious pancreatic necrosis virus VP2 subviral particles expressed in yeast. Vaccine 2007, 25, 4880–4888. [Google Scholar] [CrossRef] [PubMed]
- Norsker, N.H.; Barbosa, M.J.; Vermuë, M.H.; Wijffels, R.H. Microalgal production—A close look at the economics. Biotechnol. Adv. 2011, 29, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Acién, F.G.; Fernández, J.M.; Magán, J.J.; Molina, E. Production cost of a real microalgae production plant and strategies to reduce it. Biotechnol. Adv. 2012, 30, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Purton, S.; Baganz, F. Chitosan flocculation to aid the harvesting of the microalga Chlorella sorokiniana. Bioresour. Technol. 2013, 129, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Li, Y.; Fanning, K.; Netzel, M.; Schenk, P.M. Effect of drying, storage temperature and air exposure on astaxanthin stability from Haematococcus pluvialis. Food Res. Int. 2015, 74, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yu, Z.; Song, X.; Guan, Y. Studies on the transmission of WSSV (white spot syndrome virus) in juvenile Marsupenaeus japonicus via marine microalgae. J. Invertebr. Pathol. 2007, 95, 87–92. [Google Scholar] [CrossRef] [PubMed]
- The Convention on Biological Diversity. Nagoya Protocol. Available online: https://www.cbd.int/abs/about/ (accessed on 11 April 2018).
- Gressel, J.; van der Vlugt, C.J.B.; Bergman, H.E.N. Environmental risks of large scale cultivation of microalgae: Mitigation of spills. Algal Res. 2013, 2, 286–298. [Google Scholar] [CrossRef]
- Glass, D.J. Government regulation of the uses of genetically modified algae and other microorganisms in biofuel and bio-based chemical production. In Algal Biorefineries; Prokop, A., Bajpai, R., Zappi, M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 2, pp. 23–60. [Google Scholar]
- Henley, W.; Litaker, W.; Novoveská, L.; Duke, C.; Quemada, H.; Sayre, R.T. Initial risk assessment of genetically modified (GM) algae for commodity-scale cultivation. Algal Res. 2013, 2, 66–77. [Google Scholar] [CrossRef]
- Fu, J.; Yang, D.; Jin, M.; Liu, W.; Zhao, X.; Li, C.; Zhao, T.; Wang, J.; Gao, Z.; Shen, Z.; et al. Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: Role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol. Ecol. 2017, 26, 5318–5333. [Google Scholar] [CrossRef] [PubMed]
- Young, R.E.B.; Purton, S. Codon reassignment to facilitate genetic engineering and biocontainment in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 2016, 14, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Lucht, J.M. Public acceptance of plant biotechnology and GM crops. Viruses 2015, 7, 4254–4281. [Google Scholar] [CrossRef] [PubMed]
| Species | Site of Transgene Insertion | DNA Delivery Method and Selection System | Introduced Gene Product | Yield | Evidence for Functionality | Reference |
|---|---|---|---|---|---|---|
| Chlamydomonas reinhardtii | Chloroplast | Microparticle bombardment; restoration of photosynthesis | p57 secreted protein from Renibacterium salmoninarum, the cause of bacterial kidney disease (BKD) in salmonid fish | N.D. 1 | Induction of anti-p57 antibodies in the blood of fish fed with the dried algae | [55] |
| Microparticle bombardment; spectinomycin resistance | Viral envelope protein 28 (VP28) of white spot syndrome virus (WSSV) G-protein from infectious hematopoietic necrosis virus (IHNV) VP2 protein of infectious pancreatic necrosis virus (IPNV) p57 BKD antigen | 0.2–21% TCP 2 <0.5% TCP <0.3% TCP <0.5% TCP | N.D. N.D. N.D. N.D. | [56] | ||
| Microparticle bombardment; spectinomycin resistance | VapA and AcrV antigens of fish bacterial pathogen Aeromonas salmonicida | 0.3% TSP 3 (VapA) 0.8% TSP (AcrV) | Initial fish feeding trials revealed no adverse effects of feeding, but also no immunological response to either antigen or protection against the pathogen (see: [57]) | [58] | ||
| Agitation with glass beads; restoration of photosynthesis | VP28 of white spot syndrome virus (WSSV) | N.D. | Challenge trials showed reduced mortality from WSSV for shrimp fed with algal meal containing the VP28 antigen | [54] | ||
| Chlamydomonas reinhardtii | Nucleus | Electroporation; rescue of arginine prototroph | 14 amino acid antigenic domain of p57 fused to an endogenous plasma membrane protein | N.D. | Induction of anti-p57 antibodies in the blood of fish fed with the dried algae | [55] |
| Agitation with glass beads; paromomycin resistance | 374 bp double-stranded RNA targeting RdRp gene of yellow head virus (YHV) | 45 ng ds-RNA per 100 mL culture | Challenge trials showed 22% improvement in survival rate against YHV for shrimp fed with algal meal containing ds-RNA | [59] | ||
| Nannochloropsis oculata | Nucleus | Electroporation, fluorescence of DsRed 4 reporter | Broad spectrum antimicrobial peptide Bovine Lactoferricin (LFB) fused to DsRed | N.D. | Medaka fish fed with algal meal containing LFB showed ~85% survival against Vibrio parahymolyticus infection compared to control (~5%) | [52] |
| Dunaliella salina | Nucleus | Agitating with glass beads; phosphinothricin resistance | VP28 of white spot syndrome virus (WSSV) | 78 µg/100 mL culture | Challenge trials showed 41% survival rate of crayfish against WSSV | [53] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Charoonnart, P.; Purton, S.; Saksmerprome, V. Applications of Microalgal Biotechnology for Disease Control in Aquaculture. Biology 2018, 7, 24. https://doi.org/10.3390/biology7020024
Charoonnart P, Purton S, Saksmerprome V. Applications of Microalgal Biotechnology for Disease Control in Aquaculture. Biology. 2018; 7(2):24. https://doi.org/10.3390/biology7020024
Chicago/Turabian StyleCharoonnart, Patai, Saul Purton, and Vanvimon Saksmerprome. 2018. "Applications of Microalgal Biotechnology for Disease Control in Aquaculture" Biology 7, no. 2: 24. https://doi.org/10.3390/biology7020024





