On the Generality of Codebook Approach for Sensor-Based Human Activity Recognition
Abstract
:1. Introduction
2. Related Work
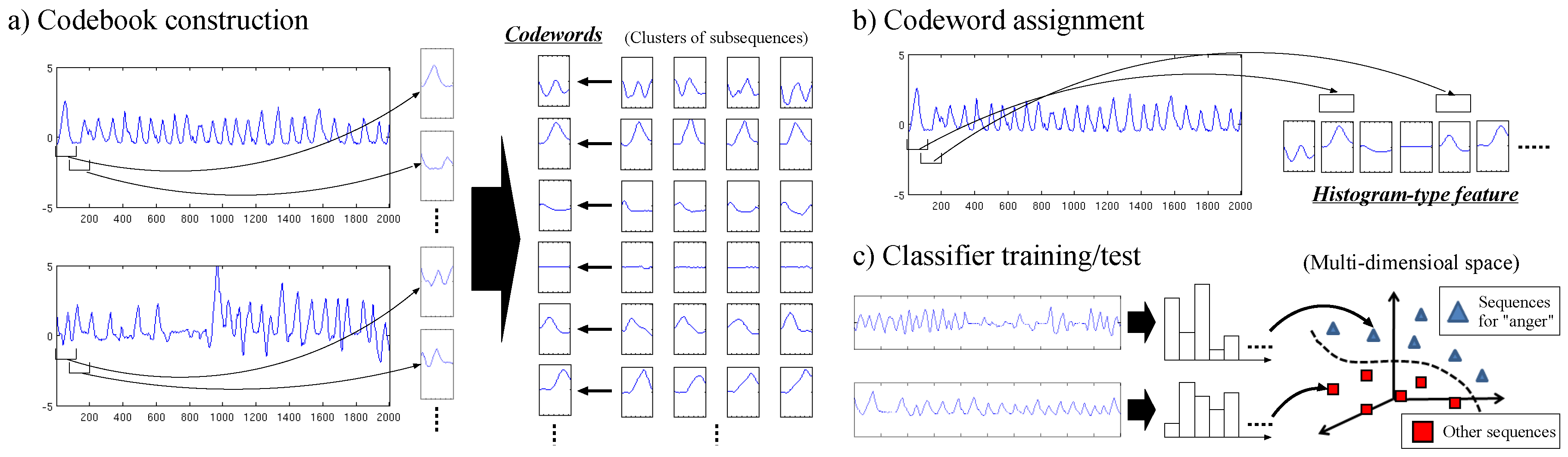
3. Codebook-Based Human Activity Recognition
3.1. Codebook Construction
3.2. Codeword Assignment
3.3. Classifier Training/Test
3.4. Fusion of Multiple Features
4. Experimental Results
4.1. Mental Activity Recognition Using Physiological Data
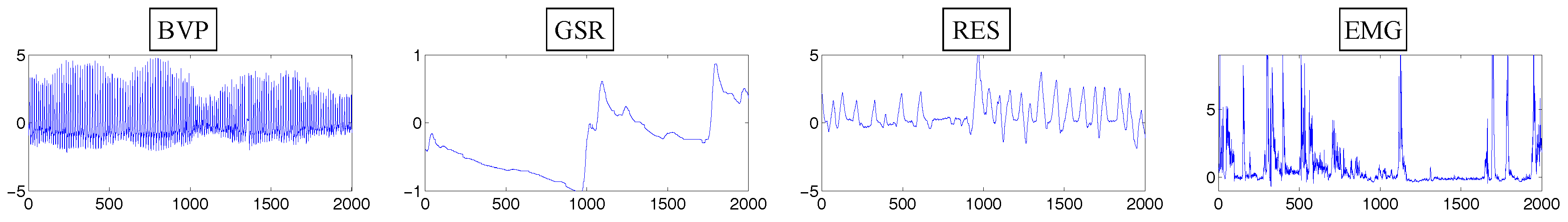
4.1.1. Dataset Description
4.1.2. Implementation Details
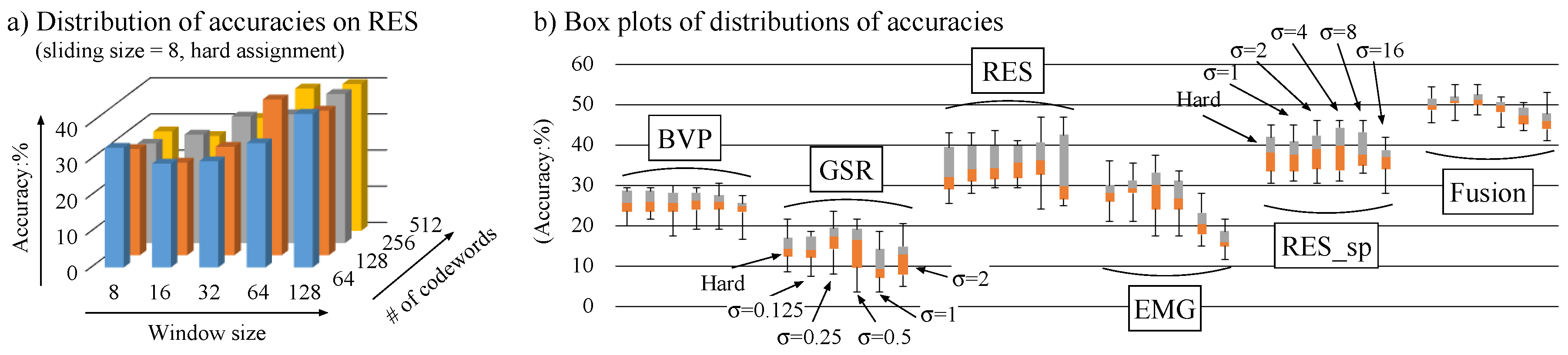
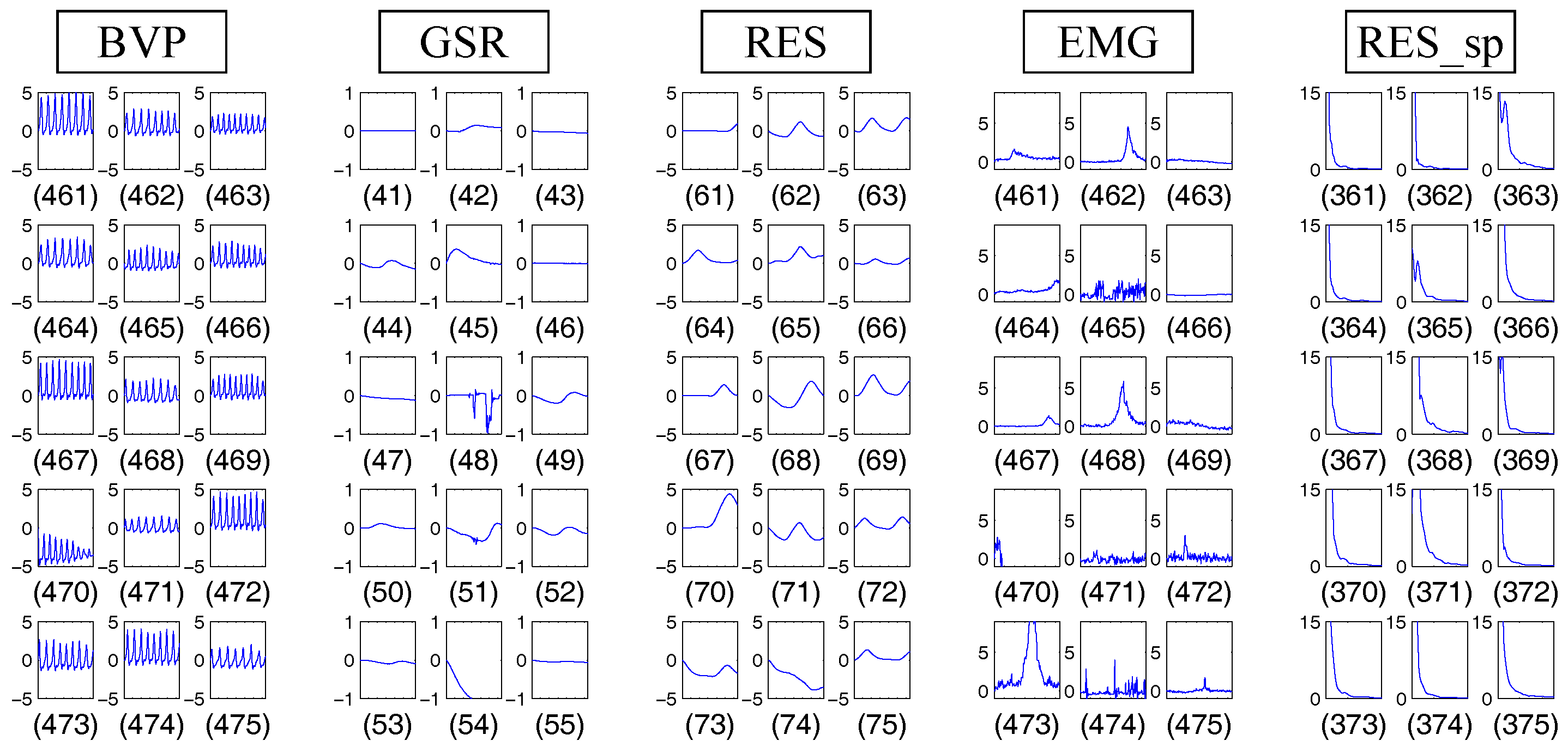
4.1.3. Results
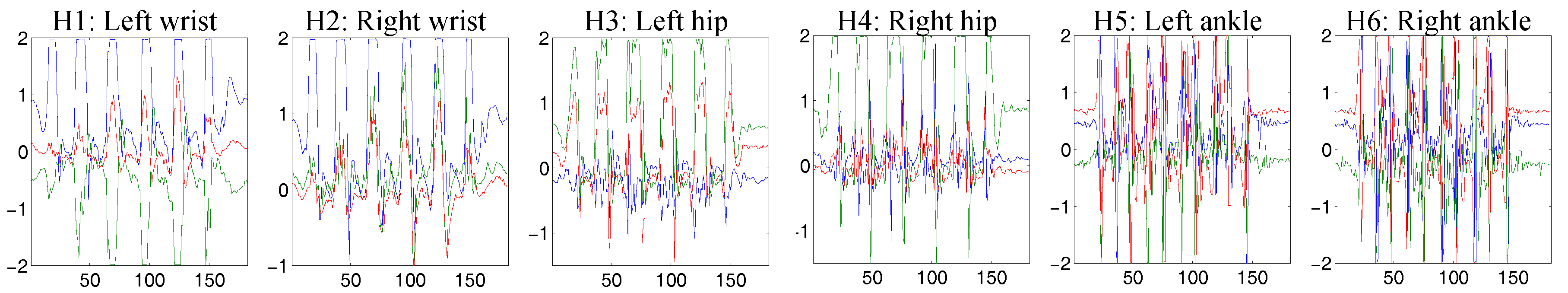
4.2. Physical Activity Recognition Using Accelerometer Data
4.2.1. Dataset Description
4.2.2. Implementation Details
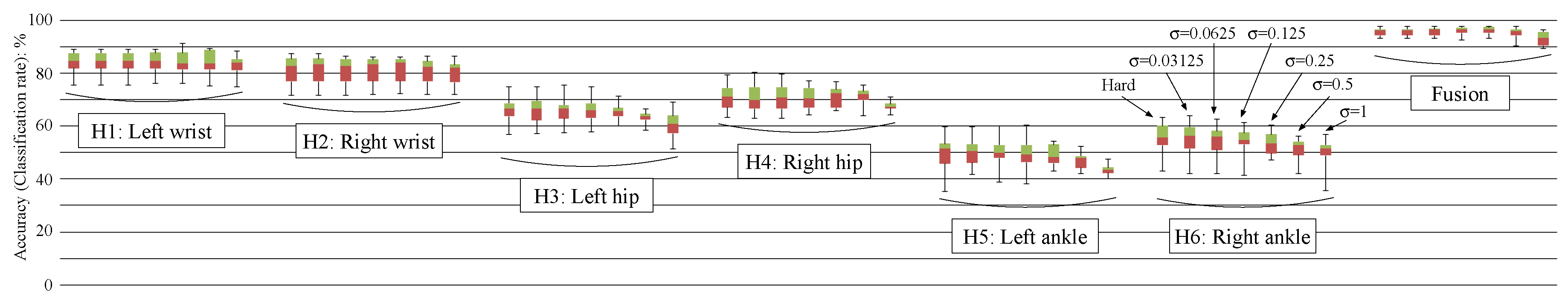
4.2.3. Results
4.3. Eye-Based Activity Recognition Using EOG Data
4.3.1. Dataset Description
4.3.2. Implementation Details
4.3.3. Results
4.4. Discussion about Computational Costs
5. Conclusions and Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| SVM | Support Vector Machine |
| BVP | Blood Volume Pressure |
| GSR | Galvanic Skin Response |
| RES | RESpiration |
| EMG | ElectroMyoGram |
| EOG | ElectroOculoGram |
| hEOG | horizontal EOG |
| vEOG | vertical EOG |
References
- Bulling, A.; Blanke, U.; Schiele, B. A Tutorial on Human Activity Recognition Using Body-worn Inertial Sensors. ACM Comput. Surv. 2014, 46, 33:1–33:33. [Google Scholar] [CrossRef]
- Chen, L.; Hoey, J.; Nugent, C.D.; Cook, D.J.; Yu, Z. Sensor-Based Activity Recognition. IEEE Trans. Syst. Man Cybern. Part C (Appl. Rev.) 2012, 42, 790–808. [Google Scholar] [CrossRef]
- Lara, D.; Labrador, A. A Survey on Human Activity Recognition using Wearable Sensors. IEEE Commun. Surv. Tutor. 2013, 15, 1192–1209. [Google Scholar] [CrossRef]
- López-Nava, H.; Muñoz-Meléndez, A. Wearable Inertial Sensors for Human Motion Analysis: A Review. IEEE Sens. J. 2016, 16, 7821–7834. [Google Scholar]
- Iosa, M.; Picerno, P.; Paolucci, S.; Morone, G. Wearable inertial sensors for human movement analysis. Expert Rev. Med. Devices 2016, 13, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Garbarino, M.; Lai, M.; Bender, D.; Picard, R.W.; Tognetti, S. Empatica E3—A Wearable Wireless Multi-sensor Device for Real-time Computerized Biofeedback and Data Acquisition. In Proceedings of the 2014 EAI 4th International Conference on Wireless Mobile Communication and Healthcare (Mobihealth), Athens, Greece, 3–5 November 2014; pp. 39–42. [Google Scholar]
- Microsoft Corporation. Microsoft Band|Official Site. Available online: www.microsoft.com/microsoft-band (accessed on 29 April 2017).
- Spire, Inc. Spire–Mindfulness & Activity Tracker. Available online: www.spire.io (accessed on 29 April 2017).
- Ishimaru, S.; Kunze, K.; Tanaka, K.; Uema, Y.; Kise, K.; Inami, M. Smart Eyewear for Interaction and Activity Recognition. In Proceedings of the CHI EA 2015—33rd Annual CHI Conference on Human Factors in Computing Systems, Seoul, Korea, 18–23 April 2015; pp. 307–310. [Google Scholar]
- Bulling, A.; Ward, A.; Gellersen, H.; Tröster, G. Eye Movement Analysis for Activity Recognition Using Electrooculography. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Picard, W.; Vyzas, E.; Healey, J. Toward Machine Emotional Intelligence: Analysis of Affective Physiological State. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 1175–1191. [Google Scholar] [CrossRef]
- Koelstra, S.; Mühl, C.; Soleymani, M.; Lee, S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. DEAP: A Database for Emotion Analysis Using Physiological Signals. IEEE Trans. Affect. Comput. 2012, 3, 18–31. [Google Scholar] [CrossRef]
- Kim, J.; Andre, E. Emotion Recognition Based on Physiological Changes in Music Listening. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 30, 2067–2083. [Google Scholar] [PubMed]
- Soleymani, M.; Lichtenauer, J.; Pun, T.; Pantic, M. A Multimodal Database for Affect Recognition and Implicit Tagging. IEEE Trans. Affect. Comput. 2012, 3, 42–55. [Google Scholar] [CrossRef]
- Hong, J.H.; Ramos, J.; Dey, A.K. Understanding Physiological Responses to Stressors During Physical Activity. In Proceedings of the 2012 ACM Conference on Ubiquitous Computing, Pittsburgh, PA, USA, 5–8 September 2012; pp. 270–279. [Google Scholar]
- Plarre, K.; Raji, A.; Hossain, S.; Ali, A.; Nakajima, M.; Al’absi, M.; Ertin, E.; Kamarck, T.; kumar, S.; Scott, M.; et al. Continuous Inference of Psychological Stress from Sensory Measurements Collected in the Natural Environment. In Proceedings of the 10th International Conference on Information Processing in Sensor Networks (IPSN), Chicago, IL, USA, 12–14 April 2011; pp. 97–108. [Google Scholar]
- Gu, T.; Wang, L.; Wu, Z.; Tao, X.; Lu, J. A Pattern Mining Approach to Sensor-Based Human Activity Recognition. IEEE Trans. Knowl. Data Eng. 2011, 23, 1359–1372. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation Learning: A Review and New Perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.; Lee, H.; Ng, A.Y. An Analysis of Single-Layer Networks in Unsupervised Feature Learning. In Proceedings of the 14th International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; pp. 215–223. [Google Scholar]
- Csurka, G.; Bray, C.; Dance, C.; Fan, L. Visual Categorization with Bags of Keypoints. In Proceedings of the ECCV International Workshop on Statistical Learning in Computer Vision, Prague, Czech Republic, 11–14 May 2004; pp. 1–22. [Google Scholar]
- Jiang, Y.G.; Yang, J.; Ngo, C.W.; Hauptmann, A.G. Representations of Keypoint-Based Semantic Concept Detection: A Comprehensive Study. IEEE Trans. Multimedia 2010, 12, 42–53. [Google Scholar] [CrossRef]
- Ofli, F.; Chaudhry, R.; Kurillo, G.; Vidal, R.; Bajcsy, R. Berkeley MHAD: A Comprehensive Multimodal Human Action Database. In Proceedings of the IEEE Workshop on Applications of Computer Vision (WACV), Tampa, FL, USA, 15–17 January 2013; pp. 53–60. [Google Scholar]
- Liu, J.; Zhong, L.; Wickramasuriya, J.; Vasudevan, V. uWave: Accelerometer-based Personalized Gesture Recognition and Its Applications. Pervasive Mob. Comput. 2009, 5, 657–675. [Google Scholar] [CrossRef]
- McGlynn, D.; Madden, M.G. An Ensemble Dynamic Time Warping Classifier with Application to Activity Recognition. In Research and Development in Intelligent Systems XXVII; Bramer, M., Petridis, M., Hopgood, A., Eds.; Springer Science & Business Media: Berlin, Germany, 2011; pp. 339–352. [Google Scholar]
- Lin, J.; Li, Y. Finding Structural Similarity in Time Series Data Using Bag-of-Patterns Representation. In Proceedings of the 21st International Conference on Scientific and Statistical Database Management, New Orleans, LA, USA, 2–4 June 2009; pp. 461–477. [Google Scholar]
- Baydogan, M.G.; Runger, G.; Tuv, E. A Bag-of-Features Framework to Classify Time Series. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2796–2802. [Google Scholar] [CrossRef] [PubMed]
- Huỳnh, T.; Blanke, U.; Schiele, B. Scalable Recognition of Daily Activities with Wearable Sensors. In Proceedings of the 3rd international conference on Location-and context-awareness, Oberpfaffenhofen, Germany, 20–21 September 2007; pp. 50–67. [Google Scholar]
- Zhang, M.; Sawchuk, A. Motion Primitive-based Human Activity Recognition Using a Bag-of-features Approach. In Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium, Miami, FL, USA, 28–30 January 2012; pp. 631–640. [Google Scholar]
- Wang, J.; Liu, P.; She, M.F.H.; Nahavandi, S.; Kouzani, A. Bag-of-words Representation for Biomedical Time Series Classification. Biomed. Signal Process. Control 2013, 8, 634–644. [Google Scholar] [CrossRef]
- Ordóñez, P.; Armstrong, T.; Oates, T.; Fackle, J. Using Modified Multivariate Bag-of-words Models to Classify Physiological Data. In Proceedings of the IEEE 11th International Conference on Data Mining Workshops (ICDMW), Vancouver, BC, Canada, 11 December 2011; pp. 534–539. [Google Scholar]
- Lin, J.; Khade, R.; Li, Y. Rotation-invariant Similarity in Time Series Using Bag-of-patterns Representation. IJIIS 2012, 39, 287–315. [Google Scholar] [CrossRef]
- Zakaria, J.; Mueen, A.; Keogh, E. Clustering Time Series Using Unsupervised-Shapelets. In Proceedings of the IEEE 12th International Conference on Data Mining, Brussels, Belgium, 10–13 December 2012; pp. 785–794. [Google Scholar]
- Martinez, H.P.; Bengio, Y.; Yannakakis, G.N. Learning Deep Physiological Models of Affect. IEEE Comput. 2013, 8, 20–33. [Google Scholar] [CrossRef]
- Yang, J.B.; Nguyen, M.N.; San, P.P.; Li, X.L.; Krishnaswamy, S. Deep Convolutional Neural Networks on Multichannel Time Series for Human Activity Recognition. In Proceedings of the 24th International Conference on Artificial Intelligence, Buenos Aires, Argentina, 25–31 July 2015; pp. 3995–4001. [Google Scholar]
- Bengio, Y. Practical Recommendations for Gradient-Based Training of Deep Architectures. In Neural Networks: Tricks of the Trade, 2nd ed.; Montavon, G., Orr, G.B., Müller, K.R., Eds.; Springer: Berlin, Germany, 2012; pp. 437–478. [Google Scholar]
- Han, J.; Kamber, M.; Pei, J. Data Mining: Concepts and Techniques, 3rd ed.; Morgan Kaufmann: Burlington, MA, USA, 2011. [Google Scholar]
- Van Gemert, J.C.; Veenman, C.J.; Smeulders, A.W.M.; Geusebroek, J.M. Visual Word Ambiguity. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; The MIT Press: London, UK, 2012. [Google Scholar]
- Vapnik, V.N. Statistical Learning Theory; Wiley-Interscience: New York, NY, USA, 1998. [Google Scholar]
- Chatfield, K.; Lempitsky, V.; Vedaldi, A.; Zisserman, A. The Devil is in the Details: An Evaluation of Recent Feature Encoding Methods. In Proceedings of the British Machine Vision Conference, Dundee, UK, 29 August–2 September 2011; pp. 76.1–76.12. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM: A Library for Support Vector Machines. ACM TIST 2011, 2, 27:1–27:27. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Know. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Shirahama, K.; Matsuoka, Y.; Uehara, K. Hybrid Negative Example Selection Using Visual and Conceptual Features. Multimed. Tools and Appl. 2014, 71, 967–989. [Google Scholar] [CrossRef]
- Zhang, J.; Marszalek, M.; Lazebnik, S.; Schmid, C. Local Features and Kernels for Classification of Texture and Object Categories: A Comprehensive Study. IJCV 2007, 73, 213–238. [Google Scholar] [CrossRef]
- Shirahama, K.; Uehara, K. Kobe University and Muroran Institute of Technology at TRECVID 2012 Semantic Indexing Task. In Proceedings of the TRECVID, Gaithersburg, MD, USA, 26–28 November 2012; pp. 239–247. [Google Scholar]
- Snoek, C.G.M.; Worring, M.; Smeulders, A.W.M. Early Versus Late Fusion in Semantic Video Analysis. In Proceedings of the 13th annual ACM international conference on Multimedia, Singapore, 6–11 November 2005; pp. 399–402. [Google Scholar]
- Nowak, E.; Jurie, F.; Triggs, B. Sampling Strategies for Bag-of-Features Image Classification. In Proceedings of the 9th European conference on Computer Vision, Graz, Austria, 7–13 May 2006; pp. 490–503. [Google Scholar]
- You Tube. Demonstration Video of Sensor-Based Human Activity Recognition. Available online: https://www.youtube.com/watch?v=sIL08IE_QLE&t=101s (accssed on 10 April 2017).
- Shirahama, K.; Grzegorzek, M. Towards Large-scale Multimedia Retrieval Enriched by Knowledge about Human Interpretation: Retrospective Survey. Multimed. Tools Appl. 2016, 75, 297–331. [Google Scholar] [CrossRef]
- Nguyen, M.H.; Torresani, L.; De la Torre, F.; Rother, C. Weakly Supervised Discriminative Localization and Classification: A Joint Learning Process. In Proceedings of the IEEE 12th International Conference on Computer Vision, Kyoto, Japan, 29 September–2 October 2009; pp. 1925–1932. [Google Scholar]
- Zhou, G.T.; Lan, T.; Vahdat, A.; Mori, G. Latent Maximum Margin Clustering. In Proceedings of the NIPS, Lake Tahoe, NV, USA, 5–10 December 2013; pp. 28–36. [Google Scholar]


| This Study | Picard et al. [11] | |
|---|---|---|
| Accuracy | 40.0–46.3% () |
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | Subject 5 | Subject 6 | Subject 7 | |
| Hard assignment | 51.9/35.2 | 71.9/69.9 | 73.6/55.7 | 32.0/40.8 | 76.4/69.8 | 70.8/56.3 | 75.0/73.0 |
| Soft assignment | 45.6/49.4 | 75.8/74.7 | 79.9/65.5 | 50.9/43.3 | 78.0/70.6 | 75.4/62.4 | 75.5/74.6 |
| Bulling et al. [10] | 76.6/69.4 | 88.3/77.8 | 83.0/72.2 | 46.6/47.9 | 59.5/46.0 | 89.2/86.9 | 93.0/81.9 |
| Subject 8 | Mean | ||||||
| Hard assignment | 53.1/48.4 | 63.1/56.1 | |||||
| Soft assignment | 53.9/50.1 | 66.9/61.3 | |||||
| Bulling et al. [10] | 72.9/81.9 | 76.1/70.5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirahama, K.; Grzegorzek, M. On the Generality of Codebook Approach for Sensor-Based Human Activity Recognition. Electronics 2017, 6, 44. https://doi.org/10.3390/electronics6020044
Shirahama K, Grzegorzek M. On the Generality of Codebook Approach for Sensor-Based Human Activity Recognition. Electronics. 2017; 6(2):44. https://doi.org/10.3390/electronics6020044
Chicago/Turabian StyleShirahama, Kimiaki, and Marcin Grzegorzek. 2017. "On the Generality of Codebook Approach for Sensor-Based Human Activity Recognition" Electronics 6, no. 2: 44. https://doi.org/10.3390/electronics6020044





